Matrix Metalloprotease-7 Mediates Nucleolar Assembly and Intra-nucleolar Cleaving p53 in Gefitinib-Resistant Cancer Stem Cells
- PMID: 33089100
- PMCID: PMC7559243
- DOI: 10.1016/j.isci.2020.101600
Matrix Metalloprotease-7 Mediates Nucleolar Assembly and Intra-nucleolar Cleaving p53 in Gefitinib-Resistant Cancer Stem Cells
Abstract
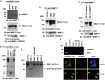
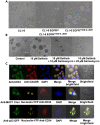
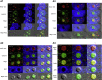
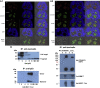
The enlarged distinct bulky-ball-like nucleolus matrix assembly is observed in most cancer stem cells (CSCs); however, the underlying mechanism is largely unknown. We show that matrix metalloproteinase-7 (MMP-7) shedding MUC-1 SEA domain releases MUC-1 C-ter, facilitating the nucleolus trafficking of p53 in gefitinib-resistant lung CSCs. The nucleolus colocalizations of p53, MUC-1 C-ter, MMP-7 and nucleolin were observed in the CD34+ CXADR+ CD44v3 + gefitinib-resistant EGFRL858R/T790M CSC colonies. MUC-1 C-ter induced a unique porous bulky-ball-shaped, cagelike nucleolus that functions as a nucleus molecular "garage" for potent tumor suppressor, p53. Nucleolus could also facilitate the novel sub-nucleus compartment for proteolytic processing p53 by MMP-7 to generate a 35 kDa fragment. Moreover, we show that salinomycin, an anti-CSC agent, disrupts nucleolus by inducing nucleoplasm translocation of p53 and sensitizing CSC to chemotherapy drugs. Thus, this study highlights the MMP-7-MUC-1-p53 axis in nucleolus as a potential therapeutic target for anti-CSCs to resolve the chemotherapy-resistance dilemma.
Keywords: Cancer; Cell Biology; Molecular Biology.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Behrendtsen O., Werb Z. Metalloproteinases regulate parietal endoderm differentiating and migrating in cultured mouse embryos. Dev. Dyn. 1997;208:255–265. - PubMed
-
- Bergers G., Coussens L.M. Extrinsic regulators of epithelial tumor progression: metalloproteinases. Curr. Opin. Genet. Dev. 2000;10:120–127. - PubMed
-
- Bharti A.K., Olson M.O., Kufe D.W., Rubin E.H. Identification of a nucleolin binding site in human topoisomerase I. J. Biol. Chem. 1996;271:1993–1997. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

