Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: an in silico approach to combat COVID-19
- PMID: 33089730
- PMCID: PMC7594184
- DOI: 10.1080/07391102.2020.1835729
Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: an in silico approach to combat COVID-19
Abstract
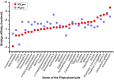
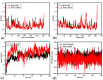
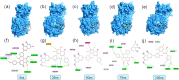
COVID-19 and its causative organism SARS-CoV2 that emerged from Wuhan city, China have paralyzed the world. With no clinically approved drugs, the global health system is struggling to find an effective treatment measure. At this crucial juncture, screening of plant-derived compounds may be an effective strategy to combat COVID-19. The present study investigated the binding affinity of phytocompounds with 3-Chymotrypsin-like (3CLpro) and Papain-like proteases (PLpro) of SARS-CoV2 using in-silico techniques. A total of 32 anti-protease phytocompounds were investigated for the binding affinity to the proteins. Docking was performed in Autodock Vina. Pharmacophore descriptors of best ligands were studied using LigandScout. Molecular dynamics (MD) simulation of apo-protein and ligand-bound complexes was carried out in YASARA software. The druglikeness properties of phytocompounds were studied using ADMETlab. Out of 32 phytochemicals, amentoflavone and gallocatechin gallate showed the best binding affinity to 3CLpro (-9.4 kcal/mol) and PLpro (-8.8 kcal/mol). Phytochemicals such as savinin, theaflavin-3,3-digallate, and kazinol-A also showed strong affinity. MD simulation revealed ligand-induced conformational changes in the protein with decreased surface area and higher stability. The RMSD/F of proteins and ligands showed stability of the protein suggesting the effective binding of the ligand in both the proteins. Both amentoflavone and gallocatechin gallate possess promising druglikeness property. The present study thus suggests that Amentoflavone and Gallocatechin gallate may be potential inhibitors of 3CLpro and PLpro proteins and effective drug candidates for SARS-CoV2. However, the findings of in silico study need to be supported by in vivo studies to establish the exact mode of action.Communicated by Ramaswamy H. Sarma.
Keywords: 3CLpro; PLpro; Phytocompounds; SARS-CoV2; docking.
Conflict of interest statement
Authors declare no conflict of interest.
Figures







Similar articles
-
Black tea bioactives as inhibitors of multiple targets of SARS-CoV-2 (3CLpro, PLpro and RdRp): a virtual screening and molecular dynamic simulation study.J Biomol Struct Dyn. 2022 Sep;40(15):7143-7166. doi: 10.1080/07391102.2021.1897679. Epub 2021 Mar 10. J Biomol Struct Dyn. 2022. PMID: 33715595
-
MD simulation and MM/PBSA identifies phytochemicals as bifunctional inhibitors of SARS-CoV-2.J Biomol Struct Dyn. 2022;40(22):12048-12061. doi: 10.1080/07391102.2021.1969285. Epub 2021 Aug 27. J Biomol Struct Dyn. 2022. PMID: 34448684
-
Identification of structural scaffold from interbioscreen (IBS) database to inhibit 3CLpro, PLpro, and RdRp of SARS-CoV-2 using molecular docking and dynamic simulation studies.J Biomol Struct Dyn. 2023;41(22):13168-13179. doi: 10.1080/07391102.2023.2175377. Epub 2023 Feb 9. J Biomol Struct Dyn. 2023. PMID: 36757134
-
Harnessing and bioprospecting botanical-based herbal medicines against potential drug targets for COVID-19: a review coupled molecular docking studies.J Biomol Struct Dyn. 2023 Apr 27:1-23. doi: 10.1080/07391102.2023.2187634. Online ahead of print. J Biomol Struct Dyn. 2023. PMID: 37105230 Review.
-
Structure-Function Characteristics of SARS-CoV-2 Proteases and Their Potential Inhibitors from Microbial Sources.Microorganisms. 2021 Nov 30;9(12):2481. doi: 10.3390/microorganisms9122481. Microorganisms. 2021. PMID: 34946083 Free PMC article. Review.
Cited by
-
Antiviral peptides against the main protease of SARS-CoV-2: A molecular docking and dynamics study.Arab J Chem. 2021 Sep;14(9):103315. doi: 10.1016/j.arabjc.2021.103315. Epub 2021 Jul 14. Arab J Chem. 2021. PMID: 34909064 Free PMC article.
-
Further quantitative in silico analysis of SARS-CoV-2 S-RBD Omicron BA.4, BA.5, BA.2.75, BQ.1, and BQ.1.1 transmissibility.Talanta. 2023 Mar 1;254:124127. doi: 10.1016/j.talanta.2022.124127. Epub 2022 Nov 23. Talanta. 2023. PMID: 36462284 Free PMC article.
-
Plant-Based Phytochemical Screening by Targeting Main Protease of SARS-CoV-2 to Design Effective Potent Inhibitors.Biology (Basel). 2021 Jun 26;10(7):589. doi: 10.3390/biology10070589. Biology (Basel). 2021. PMID: 34206970 Free PMC article.
-
Novel coronavirus disease (COVID-19) pandemic: A recent mini review.Comput Struct Biotechnol J. 2021;19:612-623. doi: 10.1016/j.csbj.2020.12.033. Epub 2020 Dec 31. Comput Struct Biotechnol J. 2021. PMID: 33398233 Free PMC article. Review.
-
Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation.J Pers Med. 2021 Sep 6;11(9):888. doi: 10.3390/jpm11090888. J Pers Med. 2021. PMID: 34575665 Free PMC article.
References
-
- Chen, C.-N., Lin, C. P. C., Huang, K.-K., Chen, W.-C., Hsieh, H.-P., Liang, P.-H., & Hsu, J. T.-A. (2005). Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3'-digallate (TF3). Evidence-Based Complementary and Alternative Medicine: ECAM, 2(2), 209–215. 10.1093/ecam/neh081 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
