Circ_0000215 Increases the Expression of CXCR2 and Promoted the Progression of Glioma Cells by Sponging miR-495-3p
- PMID: 33089764
- PMCID: PMC7586024
- DOI: 10.1177/1533033820957026
Circ_0000215 Increases the Expression of CXCR2 and Promoted the Progression of Glioma Cells by Sponging miR-495-3p
Abstract
Background: In recent years, accumulating studies have found that circular RNA (circRNA) exerts a great effect on tumor progression. Circ_0000215, a novel circRNA, remains largely unknown in terms of its effect and mechanism in glioma.
Method: Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out to detect the expressions of circ_0000215, miR-495-3p and CXCR2 in human glial cell line HEB and glioma cell lines (A172, U251, U87, SHG-44, LN-18), human glioma tissues and adjacent healthy tissues. Gain- and loss-assays of circ_0000215 were conducted. Cell proliferation ability was detected via the CCK8 assay, and cell invasion ability was examined by Transwell assay. CXCR2 expression was evaluated via RT-PCR and Western blot. Moreover, bioinformatics was applied to analyze the targeting molecules of circ_0000215 and CXCR2. Verification of the relationship between these molecules were supported through the dual-luciferase reporter gene and RNA immunocoprecipitation (RIP) assay.
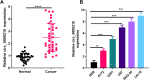
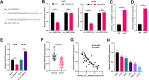
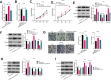
Results: Circ_0000215 and CXCR2 were remarkably upregulated in glioma tissues and cells. Overexpression of circ_0000215 notably promoted the proliferation, invasion and epithelial-mesenchymal transition (EMT) but inhibited apoptosis of glioma cells, while knocking down circ_0000215 had the opposite effects. Additionally, miR-495-3p, a sponge RNA of circ_0000215, inhibited the growth, invasion and EMT of glioma cells. Mechanistically, miR-495-3p targeted CXCR2 and negatively regulated CXCR2/PI3K/Akt pathway. However, the effects of miR-495-3p were all dampened by overexpression of circ_0000215.
Conclusion: These data demonstrated that circ_0000215 functions as a competitive endogenous RNA by sponging miR-495-3p, thus accelerating glioma progression through CXCR2 axis.
Keywords: CXCR2; circ_0000215; glioma; invasion; miR-495-3p; proliferation.
Conflict of interest statement
Figures


References
-
- Frosina G. Development of therapeutics for high grade gliomas using orthotopic rodent models. Curr Med Chem. 2013;20(26):3272–3299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

