Reference genome and demographic history of the most endangered marine mammal, the vaquita
- PMID: 33089966
- PMCID: PMC8247363
- DOI: 10.1111/1755-0998.13284
Reference genome and demographic history of the most endangered marine mammal, the vaquita
Abstract
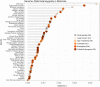
The vaquita is the most critically endangered marine mammal, with fewer than 19 remaining in the wild. First described in 1958, the vaquita has been in rapid decline for more than 20 years resulting from inadvertent deaths due to the increasing use of large-mesh gillnets. To understand the evolutionary and demographic history of the vaquita, we used combined long-read sequencing and long-range scaffolding methods with long- and short-read RNA sequencing to generate a near error-free annotated reference genome assembly from cell lines derived from a female individual. The genome assembly consists of 99.92% of the assembled sequence contained in 21 nearly gapless chromosome-length autosome scaffolds and the X-chromosome scaffold, with a scaffold N50 of 115 Mb. Genome-wide heterozygosity is the lowest (0.01%) of any mammalian species analysed to date, but heterozygosity is evenly distributed across the chromosomes, consistent with long-term small population size at genetic equilibrium, rather than low diversity resulting from a recent population bottleneck or inbreeding. Historical demography of the vaquita indicates long-term population stability at less than 5,000 (Ne) for over 200,000 years. Together, these analyses indicate that the vaquita genome has had ample opportunity to purge highly deleterious alleles and potentially maintain diversity necessary for population health.
Keywords: Conservation genomics; Phocoena sinus; Vertebrate Genomes Project; genome diversity; historical demography; porpoise.
© 2020 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Figures

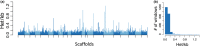

References
-
- Armstrong, E. E. , Taylor, R. W. , Prost, S. , Blinston, P. , van der Meer, E. , Madzikanda, H. , Mufute, O. , Mandisodza‐Chikerema, R. , Stuelpnagel, J. , Sillero‐Zubiri, C. , & Petrov, D. (2019). Cost‐effective assembly of the African wild dog (Lycaon pictus) genome using linked reads. Gigascience, 8(2), 10.1093/gigascience/giy124 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

