Brucella Genomics: Macro and Micro Evolution
- PMID: 33092044
- PMCID: PMC7589603
- DOI: 10.3390/ijms21207749
Brucella Genomics: Macro and Micro Evolution
Abstract
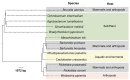
Brucella organisms are responsible for one of the most widespread bacterial zoonoses, named brucellosis. The disease affects several species of animals, including humans. One of the most intriguing aspects of the brucellae is that the various species show a ~97% similarity at the genome level. Still, the distinct Brucella species display different host preferences, zoonotic risk, and virulence. After 133 years of research, there are many aspects of the Brucella biology that remain poorly understood, such as host adaptation and virulence mechanisms. A strategy to understand these characteristics focuses on the relationship between the genomic diversity and host preference of the various Brucella species. Pseudogenization, genome reduction, single nucleotide polymorphism variation, number of tandem repeats, and mobile genetic elements are unveiled markers for host adaptation and virulence. Understanding the mechanisms of genome variability in the Brucella genus is relevant to comprehend the emergence of pathogens.
Keywords: Brucella; IS711; SNPs; brucellosis; genome reduction; pseudogene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Stackebrandt E., Murray R.G.E., Truper H.G. Proteobacteria classis nov., a Name for the Phylogenetic Taxon That Includes the “Purple Bacteria and Their Relatives”. Int. J. Syst. Bacteriol. 1988;38:321–325. doi: 10.1099/00207713-38-3-321. - DOI
-
- Moreno E. In search of a bacterial species definition. Rev. Biol. Trop. 1997;45:753–771. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

