Biological and Clinical Changes in a Pediatric Series Treated with Off-Label JAK Inhibitors
- PMID: 33092242
- PMCID: PMC7590237
- DOI: 10.3390/ijms21207767
Biological and Clinical Changes in a Pediatric Series Treated with Off-Label JAK Inhibitors
Abstract
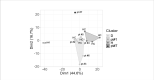
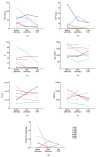
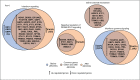
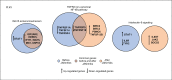
Off-label use of medications is still a common practice in pediatric rheumatology. JAK inhibitors are authorized in adults in the treatment of rheumatoid arthritis, psoriatic arthritis and ulcerative colitis. Although their use is not authorized yet in children, JAK inhibitors, based on their mechanism of action and on clinical experiences in small series, have been suggested to be useful in the treatment of pediatric interferon-mediated inflammation. Accordingly, an increased interferon score may help to identify those patients who might benefit of JAK inhibitors. We describe the clinical experience with JAK inhibitors in seven children affected with severe inflammatory conditions and we discuss the correlation between clinical features and transcriptomic data. Clinical improvements were recorded in all cases. A reduction of interferon signaling was recorded in three out of seven subjects at last follow-up, irrespectively from clinical improvements. Other signal pathways with significant differences between patients and controls included upregulation of DNA repair pathway and downregulation of extracellular collagen homeostasis. Two patients developed drug-related adverse events, which were considered serious in one case. In conclusion, JAK inhibitors may offer a valuable option for children with severe interferon-mediated inflammatory disorders reducing the interferon score as well as influencing other signal pathways that deserve future studies.
Keywords: Janus kinase inhibitors; interferon signature; juvenile idiopathic arthritis; juvenile systemic erythematosus lupus; juvenile systemic sclerosis; monogenic interferonopathies; off-label medications; pediatric rheumatology; transcriptomics.
Conflict of interest statement
Alberto Tommasini is involved in the I4V-MC-JAHV and I4V-MC-JAHU trials studying baricitinib in juvenile idiopathic arthritis sponsored by Eli Lilly corp. However, none of the patients included in the present report have been evaluated or enrolled in this study. The authors declare that Eli Lilly had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures









References
-
- Kim H., de Jesus A.A., Brooks S.R., Liu Y., Huang Y., VanTries R., Montealegre Sanchez G.A., Rotman Y., Gadina M., Goldbach-Mansky R. Development of a Validated Interferon Score Using NanoString Technology. J. Interferon Cytokine Res. 2018;38:171–185. doi: 10.1089/jir.2017.0127. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

