Genome-Wide Co-Expression Distributions as a Metric to Prioritize Genes of Functional Importance
- PMID: 33092259
- PMCID: PMC7593939
- DOI: 10.3390/genes11101231
Genome-Wide Co-Expression Distributions as a Metric to Prioritize Genes of Functional Importance
Abstract
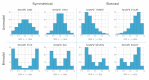
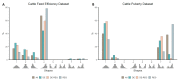
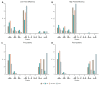
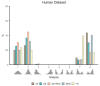
Genome-wide gene expression analysis are routinely used to gain a systems-level understanding of complex processes, including network connectivity. Network connectivity tends to be built on a small subset of extremely high co-expression signals that are deemed significant, but this overlooks the vast majority of pairwise signals. Here, we developed a computational pipeline to assign to every gene its pair-wise genome-wide co-expression distribution to one of 8 template distributions shapes varying between unimodal, bimodal, skewed, or symmetrical, representing different proportions of positive and negative correlations. We then used a hypergeometric test to determine if specific genes (regulators versus non-regulators) and properties (differentially expressed or not) are associated with a particular distribution shape. We applied our methodology to five publicly available RNA sequencing (RNA-seq) datasets from four organisms in different physiological conditions and tissues. Our results suggest that genes can be assigned consistently to pre-defined distribution shapes, regarding the enrichment of differential expression and regulatory genes, in situations involving contrasting phenotypes, time-series, or physiological baseline data. There is indeed a striking additional biological signal present in the genome-wide distribution of co-expression values which would be overlooked by currently adopted approaches. Our method can be applied to extract further information from transcriptomic data and help uncover the molecular mechanisms involved in the regulation of complex biological process and phenotypes.
Keywords: correlated gene expression; gene regulation; transcriptome analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Swami M. Networking complex traits. Nat. Rev. Genet. 2009;10:2566. doi: 10.1038/nrg2566. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

