Gene Regulation and Cellular Metabolism: An Essential Partnership
- PMID: 33092903
- PMCID: PMC7969386
- DOI: 10.1016/j.tig.2020.09.018
Gene Regulation and Cellular Metabolism: An Essential Partnership
Abstract
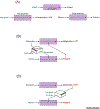
It is recognized that cell metabolism is tightly connected to other cellular processes such as regulation of gene expression. Metabolic pathways not only provide the precursor molecules necessary for gene expression, but they also provide ATP, the primary fuel driving gene expression. However, metabolic conditions are highly variable since nutrient uptake is not a uniform process. Thus, cells must continually calibrate gene expression to their changing metabolite and energy budgets. This review discusses recent advances in understanding the molecular and biophysical mechanisms that connect metabolism and gene regulation as cells navigate their growth, proliferation, and differentiation. Particular focus is given to these mechanisms in the context of organismal development.
Keywords: ATP; gene regulation; glucose metabolism.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures



References
-
- Beckwith J (2011) The operon as paradigm: normal science and the beginning of biological complexity. J Mol Biol 409 (1), 7–13. - PubMed
-
- Klar AJ and Halvorson HO (1974) Studies on the positive regulatory gene, GAL4, in regulation of galactose catabolic enzymes in Saccharomyces cerevisiae. Mol Gen Genet 135 (3), 203–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

