Metabolic Coordination of Cell Fate by α-Ketoglutarate-Dependent Dioxygenases
- PMID: 33092942
- PMCID: PMC7748998
- DOI: 10.1016/j.tcb.2020.09.010
Metabolic Coordination of Cell Fate by α-Ketoglutarate-Dependent Dioxygenases
Abstract
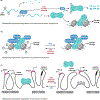
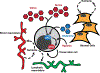
Cell fate determination requires faithful execution of gene expression programs, which are increasingly recognized to respond to metabolic inputs. In particular, the family of α-ketoglutarate (αKG)-dependent dioxygenases, which include several chromatin-modifying enzymes, are emerging as key mediators of metabolic control of cell fate. αKG-dependent dioxygenases consume the metabolite αKG (also known as 2-oxoglutarate) as an obligate cosubstrate and are inhibited by succinate, fumarate, and 2-hydroxyglutarate. Here, we review the role of these metabolites in the control of dioxygenase activity and cell fate programs. We discuss the biochemical and transcriptional mechanisms enabling these metabolites to control cell fate and review evidence that nutrient availability shapes tissue-specific fate programs via αKG-dependent dioxygenases.
Keywords: 2-hydroxyglutarate; alpha-ketoglutarate; cell fate; chromatin modifications; succinate; αKG-dependent dioxygenases.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

