SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis
- PMID: 33093148
- PMCID: PMC7721335
- DOI: 10.1105/tpc.20.00283
SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis
Abstract
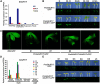
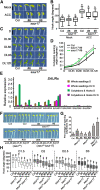
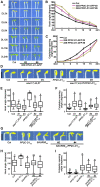
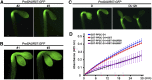
Following germination in the dark, Arabidopsis (Arabidopsis thaliana) seedlings undergo etiolation and develop apical hooks, closed cotyledons, and rapidly elongating hypocotyls. Upon light perception, the seedlings de-etiolate, which includes the opening of apical hooks and cotyledons. Here, we identify Arabidopsis Small Auxin Up RNA17 (SAUR17) as a downstream effector of etiolation, which serves to bring about apical hook formation and closed cotyledons. SAUR17 is highly expressed in apical hooks and cotyledons and is repressed by light. The apical organs also express a group of light-inducing SAURs, as represented by SAUR50, which promote hook and cotyledon opening. The development of etiolated or de-etiolated apical structures requires asymmetric differential cell growth. We present evidence that the opposing actions of SAUR17 and SAUR50 on apical development largely result from their antagonistic regulation of Protein Phosphatase 2C D-clade 1 (PP2C-D1), a phosphatase that suppresses cell expansion and promotes apical hook development in the dark. SAUR50 inhibits PP2C-D1, whereas SAUR17 has a higher affinity for PP2C-D1 without inhibiting its activity. PP2C-D1 predominantly associates with SAUR17 in etiolated seedlings, which shields it from inhibitory SAURs such as SAUR50. Light signals turn off SAUR17 and upregulate a subgroup of SAURs including SAUR50 at the inner side of the hook and cotyledon cells, leading to cell expansion and unfolding of the hook and cotyledons.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures





Similar articles
-
The Transcription Factors TCP4 and PIF3 Antagonistically Regulate Organ-Specific Light Induction of SAUR Genes to Modulate Cotyledon Opening during De-Etiolation in Arabidopsis.Plant Cell. 2019 May;31(5):1155-1170. doi: 10.1105/tpc.18.00803. Epub 2019 Mar 25. Plant Cell. 2019. PMID: 30914467 Free PMC article.
-
APYRASE1/2 mediate red light-induced de-etiolation growth in Arabidopsis seedlings.Plant Physiol. 2022 Jun 27;189(3):1728-1740. doi: 10.1093/plphys/kiac150. Plant Physiol. 2022. PMID: 35357495 Free PMC article.
-
Arabidopsis SAURs are critical for differential light regulation of the development of various organs.Proc Natl Acad Sci U S A. 2016 May 24;113(21):6071-6. doi: 10.1073/pnas.1604782113. Epub 2016 Apr 26. Proc Natl Acad Sci U S A. 2016. PMID: 27118848 Free PMC article.
-
[The molecular mechanism of apical hook development in dicot plant].Yi Chuan. 2021 Aug 20;43(8):723-736. doi: 10.16288/j.yczz.21-105. Yi Chuan. 2021. PMID: 34413013 Review. Chinese.
-
Strategies of seedlings to overcome their sessile nature: auxin in mobility control.Front Plant Sci. 2015 Apr 14;6:218. doi: 10.3389/fpls.2015.00218. eCollection 2015. Front Plant Sci. 2015. PMID: 25926839 Free PMC article. Review.
Cited by
-
Integrated Transcriptomic and Metabolomic Analysis Reveals the Mechanism of Gibberellic acid Regulates the Growth and Flavonoid Synthesis in Phellodendron chinense Schneid Seedlings.Int J Mol Sci. 2023 Nov 7;24(22):16045. doi: 10.3390/ijms242216045. Int J Mol Sci. 2023. PMID: 38003235 Free PMC article.
-
Brassinosteroids promote etiolated apical structures in darkness by amplifying the ethylene response via the EBF-EIN3/PIF3 circuit.Plant Cell. 2023 Jan 2;35(1):390-408. doi: 10.1093/plcell/koac316. Plant Cell. 2023. PMID: 36321994 Free PMC article.
-
Molecular Processes of Dodder Haustorium Formation on Host Plant under Low Red/Far Red (R/FR) Irradiation.Int J Mol Sci. 2022 Jul 7;23(14):7528. doi: 10.3390/ijms23147528. Int J Mol Sci. 2022. PMID: 35886875 Free PMC article.
-
Overexpression of PagSTOMAGEN, a Positive Regulator of Stomatal Density, Promotes Vegetative Growth in Poplar.Int J Mol Sci. 2022 Sep 5;23(17):10165. doi: 10.3390/ijms231710165. Int J Mol Sci. 2022. PMID: 36077563 Free PMC article.
-
Genome-Wide Identification of the SAUR Gene Family in Wax Gourd (Benincasa hispida) and Functional Characterization of BhSAUR60 during Fruit Development.Int J Mol Sci. 2022 Nov 14;23(22):14021. doi: 10.3390/ijms232214021. Int J Mol Sci. 2022. PMID: 36430500 Free PMC article.
References
-
- Atamian H.S., Creux N.M., Brown E.A., Garner A.G., Blackman B.K., Harmer S.L.(2016). Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 353: 587–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

