Visualization of the HIV-1 Env glycan shield across scales
- PMID: 33093196
- PMCID: PMC7668054
- DOI: 10.1073/pnas.2000260117
Visualization of the HIV-1 Env glycan shield across scales
Abstract
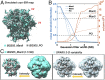
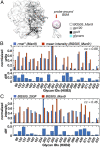
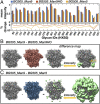
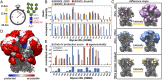
The dense array of N-linked glycans on the HIV-1 envelope glycoprotein (Env), known as the "glycan shield," is a key determinant of immunogenicity, yet intrinsic heterogeneity confounds typical structure-function analysis. Here, we present an integrated approach of single-particle electron cryomicroscopy (cryo-EM), computational modeling, and site-specific mass spectrometry (MS) to probe glycan shield structure and behavior at multiple levels. We found that dynamics lead to an extensive network of interglycan interactions that drive the formation of higher-order structure within the glycan shield. This structure defines diffuse boundaries between buried and exposed protein surface and creates a mapping of potentially immunogenic sites on Env. Analysis of Env expressed in different cell lines revealed how cryo-EM can detect subtle changes in glycan occupancy, composition, and dynamics that impact glycan shield structure and epitope accessibility. Importantly, this identified unforeseen changes in the glycan shield of Env obtained from expression in the same cell line used for vaccine production. Finally, by capturing the enzymatic deglycosylation of Env in a time-resolved manner, we found that highly connected glycan clusters are resistant to digestion and help stabilize the prefusion trimer, suggesting the glycan shield may function beyond immune evasion.
Keywords: HIV-1; cryo-EM; glycoprotein; molecular modeling; vaccine.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures






References
-
- Fenouillet E., Gluckman J. C., Jones I. M., Functions of HIV envelope glycans. Trends Biochem. Sci. 19, 65–70 (1994). - PubMed
-
- Grundner C., et al. , Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology 330, 233–248 (2004). - PubMed
-
- Lasky L. A., et al. , Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233, 209–212 (1986). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

