Ensemble transfer learning for the prediction of anti-cancer drug response
- PMID: 33093487
- PMCID: PMC7581765
- DOI: 10.1038/s41598-020-74921-0
Ensemble transfer learning for the prediction of anti-cancer drug response
Abstract
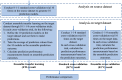
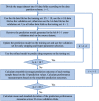
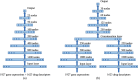
Transfer learning, which transfers patterns learned on a source dataset to a related target dataset for constructing prediction models, has been shown effective in many applications. In this paper, we investigate whether transfer learning can be used to improve the performance of anti-cancer drug response prediction models. Previous transfer learning studies for drug response prediction focused on building models to predict the response of tumor cells to a specific drug treatment. We target the more challenging task of building general prediction models that can make predictions for both new tumor cells and new drugs. Uniquely, we investigate the power of transfer learning for three drug response prediction applications including drug repurposing, precision oncology, and new drug development, through different data partition schemes in cross-validation. We extend the classic transfer learning framework through ensemble and demonstrate its general utility with three representative prediction algorithms including a gradient boosting model and two deep neural networks. The ensemble transfer learning framework is tested on benchmark in vitro drug screening datasets. The results demonstrate that our framework broadly improves the prediction performance in all three drug response prediction applications with all three prediction algorithms.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
DeepDRK: a deep learning framework for drug repurposing through kernel-based multi-omics integration.Brief Bioinform. 2021 Sep 2;22(5):bbab048. doi: 10.1093/bib/bbab048. Brief Bioinform. 2021. PMID: 33822890
-
Ensemble machine learning model trained on a new synthesized dataset generalizes well for stress prediction using wearable devices.J Biomed Inform. 2023 Dec;148:104556. doi: 10.1016/j.jbi.2023.104556. Epub 2023 Dec 2. J Biomed Inform. 2023. PMID: 38048895
-
DeepSeq2Drug: An expandable ensemble end-to-end anti-viral drug repurposing benchmark framework by multi-modal embeddings and transfer learning.Comput Biol Med. 2024 Jun;175:108487. doi: 10.1016/j.compbiomed.2024.108487. Epub 2024 Apr 17. Comput Biol Med. 2024. PMID: 38653064
-
Review of machine learning and deep learning models for toxicity prediction.Exp Biol Med (Maywood). 2023 Nov;248(21):1952-1973. doi: 10.1177/15353702231209421. Epub 2023 Dec 6. Exp Biol Med (Maywood). 2023. PMID: 38057999 Free PMC article. Review.
-
Advancing Peptide-Based Cancer Therapy with AI: In-Depth Analysis of State-of-the-Art AI Models.J Chem Inf Model. 2024 Jul 8;64(13):4941-4957. doi: 10.1021/acs.jcim.4c00295. Epub 2024 Jun 14. J Chem Inf Model. 2024. PMID: 38874445 Review.
Cited by
-
NeRD: a multichannel neural network to predict cellular response of drugs by integrating multidimensional data.BMC Med. 2022 Oct 17;20(1):368. doi: 10.1186/s12916-022-02549-0. BMC Med. 2022. PMID: 36244991 Free PMC article.
-
High dimensional predictions of suicide risk in 4.2 million US Veterans using ensemble transfer learning.Sci Rep. 2024 Jan 20;14(1):1793. doi: 10.1038/s41598-024-51762-9. Sci Rep. 2024. PMID: 38245528 Free PMC article.
-
Data augmentation and multimodal learning for predicting drug response in patient-derived xenografts from gene expressions and histology images.Front Med (Lausanne). 2023 Mar 7;10:1058919. doi: 10.3389/fmed.2023.1058919. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36960342 Free PMC article.
-
Exploring approaches for predictive cancer patient digital twins: Opportunities for collaboration and innovation.Front Digit Health. 2022 Oct 6;4:1007784. doi: 10.3389/fdgth.2022.1007784. eCollection 2022. Front Digit Health. 2022. PMID: 36274654 Free PMC article.
-
Development and validation of an ensemble learning risk model for sepsis after abdominal surgery.Arch Med Sci. 2024 Jun 6;21(1):138-152. doi: 10.5114/aoms/189505. eCollection 2025. Arch Med Sci. 2024. PMID: 40190318 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

