Neuroepithelial cell competition triggers loss of cellular juvenescence
- PMID: 33093561
- PMCID: PMC7582913
- DOI: 10.1038/s41598-020-74874-4
Neuroepithelial cell competition triggers loss of cellular juvenescence
Abstract
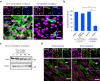
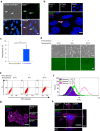
Cell competition is a cell-cell interaction mechanism which maintains tissue homeostasis through selective elimination of unfit cells. During early brain development, cells are eliminated through apoptosis. How cells are selected to undergo elimination remains unclear. Here we aimed to identify a role for cell competition in the elimination of suboptimal cells using an in vitro neuroepithelial model. Cell competition was observed when neural progenitor HypoE-N1 cells expressing RASV12 were surrounded by normal cells in the co-culture. The elimination through apoptosis was observed by cellular changes of RASV12 cells with rounding/fragmented morphology, by SYTOX blue-positivity, and by expression of apoptotic markers active caspase-3 and cleaved PARP. In this model, expression of juvenility-associated genes Srsf7 and Ezh2 were suppressed under cell-competitive conditions. Srsf7 depletion led to loss of cellular juvenescence characterized by suppression of Ezh2, cell growth impairment and enhancement of senescence-associated proteins. The cell bodies of eliminated cells were engulfed by the surrounding cells through phagocytosis. Our data indicates that neuroepithelial cell competition may have an important role for maintaining homeostasis in the neuroepithelium by eliminating suboptimal cells through loss of cellular juvenescence.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

