Epigenetic alterations in skin homing CD4+CLA+ T cells of atopic dermatitis patients
- PMID: 33093567
- PMCID: PMC7582180
- DOI: 10.1038/s41598-020-74798-z
Epigenetic alterations in skin homing CD4+CLA+ T cells of atopic dermatitis patients
Abstract
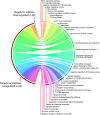
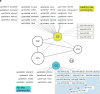
T cells expressing the cutaneous lymphocyte antigen (CLA) mediate pathogenic inflammation in atopic dermatitis (AD). The molecular alterations contributing to their dysregulation remain unclear. With the aim to elucidate putative altered pathways in AD we profiled DNA methylation levels and miRNA expression in sorted T cell populations (CD4+, CD4+CD45RA+ naïve, CD4+CLA+, and CD8+) from adult AD patients and healthy controls (HC). Skin homing CD4+CLA+ T cells from AD patients showed significant differences in DNA methylation in 40 genes compared to HC (p < 0.05). Reduced DNA methylation levels in the upstream region of the interleukin-13 gene (IL13) in CD4+CLA+ T cells from AD patients correlated with increased IL13 mRNA expression in these cells. Sixteen miRNAs showed differential expression in CD4+CLA+ T cells from AD patients targeting genes in 202 biological processes (p < 0.05). An integrated network analysis of miRNAs and CpG sites identified two communities of strongly interconnected regulatory elements with strong antagonistic behaviours that recapitulated the differences between AD patients and HC. Functional analysis of the genes linked to these communities revealed their association with key cytokine signaling pathways, MAP kinase signaling and protein ubiquitination. Our findings support that epigenetic mechanisms play a role in the pathogenesis of AD by affecting inflammatory signaling molecules in skin homing CD4+CLA+ T cells and uncover putative molecules participating in AD pathways.
Conflict of interest statement
A. Scheynius is a member in the Joint Steering Committee for the Human Translational Microbiome Program at Karolinska Institutet together with Ferring Pharmaceuticals, Switzerland. The rest of the authors declare that they have no relevant conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

