Messenger RNA enrichment using synthetic oligo(T) click nucleic acids
- PMID: 33094748
- PMCID: PMC7891491
- DOI: 10.1039/d0cc05815g
Messenger RNA enrichment using synthetic oligo(T) click nucleic acids
Abstract
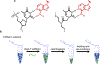
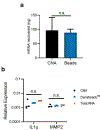
Enrichment of mRNA is a key step in a number of molecular biology techniques, particularly in the rapidly growing field of transcriptomics. Currently, mRNA is isolated using oligo(thymine) DNA (oligo(dT)) immobilized on solid supports, which binds to the poly(A) tail of mRNA to pull the mRNA out of solution through the use of magnets or centrifugal filters. Here, a simple method to isolate mRNA by complexing it with synthetic click nucleic acids (CNAs) is described. Oligo(T) CNA bound efficiently to mRNA, and because of the insolubility of CNA in water, >90% of mRNA was readily removed from solution using this method. Simple washing, buffer exchange, and heating steps enabled mRNA's enrichment from total RNA, with a yield of 3.1 ± 1.5% of the input total RNA by mass, comparable to the yield from commercially available mRNA enrichment beads. Further, the integrity and activity of mRNA after CNA-facilitated pulldown and release was evaluated through two assays. In vitro translation of EGFP mRNA confirmed the translatability of mRNA into functional protein and RT-qPCR was used to amplify enriched mRNA from total RNA extracts and compare gene expression to results obtained using commercially available products.
Conflict of interest statement
Conflicts of interest
The authors declare no conflicts of interest.
Figures
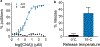

Similar articles
-
Click Nucleic Acid-DNA Binding Behavior: Dependence on Length, Sequence, and Ionic Strength.Biomacromolecules. 2020 Oct 12;21(10):4205-4211. doi: 10.1021/acs.biomac.0c00996. Epub 2020 Sep 25. Biomacromolecules. 2020. PMID: 32915548
-
Direct isolation of poly(A)+ RNA from 4 M guanidine thiocyanate-lysed cell extracts using locked nucleic acid-oligo(T) capture.Nucleic Acids Res. 2004 Apr 19;32(7):e64. doi: 10.1093/nar/gnh056. Nucleic Acids Res. 2004. PMID: 15096560 Free PMC article.
-
Isolation of Poly(A)+ Messenger RNA Using Magnetic Oligo(dT) Beads.Cold Spring Harb Protoc. 2019 Oct 1;2019(10). doi: 10.1101/pdb.prot101733. Cold Spring Harb Protoc. 2019. PMID: 31575797
-
Localization in situ of specific mRNA using thymine-thymine dimerized DNA probes. Sensitive and reliable non-radioactive in situ hybridization.Acta Pathol Jpn. 1990 Nov;40(11):793-807. doi: 10.1111/j.1440-1827.1990.tb02492.x. Acta Pathol Jpn. 1990. PMID: 2077813 Review.
-
The dynamic machinery of mRNA elongation.Curr Opin Struct Biol. 2005 Apr;15(2):197-203. doi: 10.1016/j.sbi.2005.03.002. Curr Opin Struct Biol. 2005. PMID: 15837179 Review.
Cited by
-
Challenges and Perspective in Integrated Multi-Omics in Gut Microbiota Studies.Biomolecules. 2021 Feb 17;11(2):300. doi: 10.3390/biom11020300. Biomolecules. 2021. PMID: 33671370 Free PMC article. Review.
-
Epitranscriptomics in parasitic protists: Role of RNA chemical modifications in posttranscriptional gene regulation.PLoS Pathog. 2022 Dec 22;18(12):e1010972. doi: 10.1371/journal.ppat.1010972. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36548245 Free PMC article. Review.
-
Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer.Cancer Med. 2021 Dec;10(24):8725-8740. doi: 10.1002/cam4.4398. Epub 2021 Nov 18. Cancer Med. 2021. PMID: 34796685 Free PMC article. Review.
-
Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.).Int J Mol Sci. 2022 Oct 26;23(21):12981. doi: 10.3390/ijms232112981. Int J Mol Sci. 2022. PMID: 36361778 Free PMC article.
-
The Regulation of Ferroptosis by Noncoding RNAs.Int J Mol Sci. 2023 Aug 28;24(17):13336. doi: 10.3390/ijms241713336. Int J Mol Sci. 2023. PMID: 37686142 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

