Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses
- PMID: 33096040
- PMCID: PMC7772752
- DOI: 10.1016/j.immuni.2020.10.005
Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses
Abstract
Polyreactivity is the ability of a single antibody to bind to multiple molecularly distinct antigens and is a common feature of antibodies induced upon pathogen exposure. However, little is known about the role of polyreactivity during anti-influenza virus antibody responses. By analyzing more than 500 monoclonal antibodies (mAbs) derived from B cells induced by numerous influenza virus vaccines and infections, we found mAbs targeting conserved neutralizing influenza virus hemagglutinin epitopes were polyreactive. Polyreactive mAbs were preferentially induced by novel viral exposures due to their broad viral binding breadth. Polyreactivity augmented mAb viral binding strength by increasing antibody flexibility, allowing for adaption to imperfectly conserved epitopes. Lastly, we found affinity-matured polyreactive B cells were typically derived from germline polyreactive B cells that were preferentially selected to participate in B cell responses over time. Together, our data reveal that polyreactivity is a beneficial feature of antibodies targeting conserved epitopes.
Keywords: antibody flexibility; broadly neutralizing antibodies; influenza viruses; monoclonal antibodies; polyreactivity.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The Icahn School of Medicine at Mount Sinai has submitted patent applications on universal influenza virus vaccines naming R.N., A.G.-S., P.P., and F.K. as inventors.
Figures
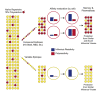
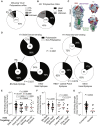

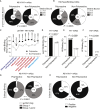

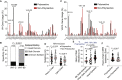

References
-
- Adelman S.A., Doll J.D. Generalized Langevin equation approach for atom/solid-surface scattering: Inelastic studies. J. Chem. Phys. 1975;64:2375.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

