Significant compaction of H4 histone tail upon charge neutralization by acetylation and its mimics, possible effects on chromatin structure
- PMID: 33096105
- PMCID: PMC8608375
- DOI: 10.1016/j.jmb.2020.10.017
Significant compaction of H4 histone tail upon charge neutralization by acetylation and its mimics, possible effects on chromatin structure
Abstract
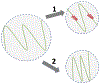
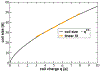
The intrinsically disordered, positively charged H4 histone tail is important for chromatin structure and function. We have explored conformational ensembles of human H4 tail in solution, with varying levels of charge neutralization via acetylation or amino-acid substitutions such as K→Q. We have employed an explicit water model shown recently to be well suited for simulations of intrinsically disordered proteins. Upon progressive neutralization of the H4, its radius of gyration decreases linearly with the tail charge q, the trend is explained using a simple polymer model. While the wild type state (q=+8) is essentially a random coil, hyper-acetylated H4 (q=+3) is virtually as compact and stable as a globular protein of the same number of amino-acids. Conformational ensembles of acetylated H4 match the corresponding K→X substitutions only approximately: based on the ensemble similarity, we propose K→M as a possible alternative to the commonly used K→Q. Possible effects of the H4 tail compaction on chromatin structure are discussed within a qualitative model in which the chromatin is highly heterogeneous, easily inter-converting between various structural forms. We predict that upon progressive charge neutralization of the H4 tail, the least compact sub-states of chromatin de-condense first, followed by de-condensation of more compact structures, e.g. those that harbor a high fraction of stacked di-nucleosomes. The predicted hierarchy of DNA accessibility increase upon progressive acetylation of H4 might be utilized by the cell for selective DNA accessibility control.
Keywords: Acetylation; Chromatin compaction; DNA accessibility; Intrinsically disordered.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
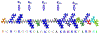
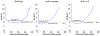
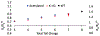
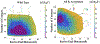

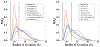
Similar articles
-
Comprehensive structural analysis of mutant nucleosomes containing lysine to glutamine (KQ) substitutions in the H3 and H4 histone-fold domains.Biochemistry. 2011 Sep 13;50(36):7822-32. doi: 10.1021/bi201021h. Epub 2011 Aug 17. Biochemistry. 2011. PMID: 21812398
-
Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle.J Mol Biol. 2021 Mar 19;433(6):166881. doi: 10.1016/j.jmb.2021.166881. Epub 2021 Feb 20. J Mol Biol. 2021. PMID: 33617899 Free PMC article.
-
Nucleosome Histone Tail Conformation and Dynamics: Impacts of Lysine Acetylation and a Nearby Minor Groove Benzo[a]pyrene-Derived Lesion.Biochemistry. 2017 Apr 11;56(14):1963-1973. doi: 10.1021/acs.biochem.6b01208. Epub 2017 Mar 22. Biochemistry. 2017. PMID: 28304160 Free PMC article.
-
Breaths, Twists, and Turns of Atomistic Nucleosomes.J Mol Biol. 2021 Mar 19;433(6):166744. doi: 10.1016/j.jmb.2020.166744. Epub 2020 Dec 10. J Mol Biol. 2021. PMID: 33309853 Review.
-
Secondary structures of the core histone N-terminal tails: their role in regulating chromatin structure.Subcell Biochem. 2013;61:37-55. doi: 10.1007/978-94-007-4525-4_2. Subcell Biochem. 2013. PMID: 23150245 Review.
Cited by
-
Impacts of Nucleosome Positioning Elements and Pre-Assembled Chromatin States on Expression and Retention of Transgenes.Genes (Basel). 2024 Sep 21;15(9):1232. doi: 10.3390/genes15091232. Genes (Basel). 2024. PMID: 39336823 Free PMC article.
-
Binding of regulatory proteins to nucleosomes is modulated by dynamic histone tails.Nat Commun. 2021 Sep 6;12(1):5280. doi: 10.1038/s41467-021-25568-6. Nat Commun. 2021. PMID: 34489435 Free PMC article.
-
Conformational Dynamics of the Nucleosomal Histone H2B Tails Revealed by Molecular Dynamics Simulations.J Chem Inf Model. 2024 Jun 24;64(12):4709-4726. doi: 10.1021/acs.jcim.4c00059. Epub 2024 Jun 12. J Chem Inf Model. 2024. PMID: 38865599 Free PMC article.
-
Acetylation-Dependent Compaction of the Histone H4 Tail Ensemble.J Phys Chem B. 2024 Oct 31;128(43):10636-10649. doi: 10.1021/acs.jpcb.4c05701. Epub 2024 Oct 22. J Phys Chem B. 2024. PMID: 39437158 Free PMC article.
-
The epigenetic mechanisms involved in the treatment resistance of glioblastoma.Cancer Drug Resist. 2025 Mar 13;8:12. doi: 10.20517/cdr.2024.157. eCollection 2025. Cancer Drug Resist. 2025. PMID: 40201311 Free PMC article. Review.
References
-
- Henikoff S, Nucleosome destabilization in the epigenetic regulation of gene expression., Nat Rev Genet 9 (2008) 15–26. - PubMed
-
- Olins A, Olins D, Spheroid chromatin units (v bodies), Science 183 (1974) 330–332. - PubMed
-
- Woodcock C, Ultrastructure of inactive chromatin, J.Cell.Biol 59 (1973) A368.
-
- Kornberg R, Chromatin structure: A repeating unit of histones and DNA, Science 184 (1974) 868–871. - PubMed
-
- Onufriev AV, Schiessel H, The nucleosome: from structure to function through physics, Current Opinion in Structural Biology 56 (2019) 119–130. Sequences and Topology Carbohydrates. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

