Cutting the Brakes on Ras-Cytoplasmic GAPs as Targets of Inactivation in Cancer
- PMID: 33096593
- PMCID: PMC7588890
- DOI: 10.3390/cancers12103066
Cutting the Brakes on Ras-Cytoplasmic GAPs as Targets of Inactivation in Cancer
Abstract
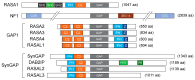
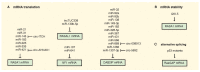
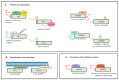
The Ras pathway is frequently deregulated in cancer, actively contributing to tumor development and progression. Oncogenic activation of the Ras pathway is commonly due to point mutation of one of the three Ras genes, which occurs in almost one third of human cancers. In the absence of Ras mutation, the pathway is frequently activated by alternative means, including the loss of function of Ras inhibitors. Among Ras inhibitors, the GTPase-Activating Proteins (RasGAPs) are major players, given their ability to modulate multiple cancer-related pathways. In fact, most RasGAPs also have a multi-domain structure that allows them to act as scaffold or adaptor proteins, affecting additional oncogenic cascades. In cancer cells, various mechanisms can cause the loss of function of Ras inhibitors; here, we review the available evidence of RasGAP inactivation in cancer, with a specific focus on the mechanisms. We also consider extracellular inputs that can affect RasGAP levels and functions, implicating that specific conditions in the tumor microenvironment can foster or counteract Ras signaling through negative or positive modulation of RasGAPs. A better understanding of these conditions might have relevant clinical repercussions, since treatments to restore or enhance the function of RasGAPs in cancer would help circumvent the intrinsic difficulty of directly targeting the Ras protein.
Keywords: GTPase-Activating Proteins; RAS oncogene; cell signaling; mechanisms of transformation; signal transduction; tumor suppressor genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A Systematic Analysis of Expression and Function of RAS GTPase-Activating Proteins (RASGAPs) in Urological Cancers: A Mini-Review.Cancers (Basel). 2025 Apr 28;17(9):1485. doi: 10.3390/cancers17091485. Cancers (Basel). 2025. PMID: 40361412 Free PMC article. Review.
-
Pumping the brakes on RAS - negative regulators and death effectors of RAS.J Cell Sci. 2020 Feb 10;133(3):jcs238865. doi: 10.1242/jcs.238865. J Cell Sci. 2020. PMID: 32041893 Free PMC article. Review.
-
Tissue-specific functions of the Caenorhabditis elegans p120 Ras GTPase activating protein GAP-3.Dev Biol. 2008 Nov 15;323(2):166-76. doi: 10.1016/j.ydbio.2008.08.026. Epub 2008 Sep 5. Dev Biol. 2008. PMID: 18805410
-
Ras-Specific GTPase-Activating Proteins-Structures, Mechanisms, and Interactions.Cold Spring Harb Perspect Med. 2019 Mar 1;9(3):a031500. doi: 10.1101/cshperspect.a031500. Cold Spring Harb Perspect Med. 2019. PMID: 30104198 Free PMC article. Review.
-
Differential Regulation of RasGAPs in Cancer.Genes Cancer. 2011 Mar;2(3):288-97. doi: 10.1177/1947601911407330. Genes Cancer. 2011. PMID: 21779499 Free PMC article.
Cited by
-
An update on the tumor-suppressive functions of the RasGAP protein DAB2IP with focus on therapeutic implications.Cell Death Differ. 2024 Jul;31(7):844-854. doi: 10.1038/s41418-024-01332-3. Epub 2024 Jun 20. Cell Death Differ. 2024. PMID: 38902547 Free PMC article. Review.
-
The Tumor Suppressor DAB2IP Is Regulated by Cell Contact and Contributes to YAP/TAZ Inhibition in Confluent Cells.Cancers (Basel). 2023 Jun 27;15(13):3379. doi: 10.3390/cancers15133379. Cancers (Basel). 2023. PMID: 37444489 Free PMC article.
-
Atomistic simulations reveal impacts of missense mutations on the structure and function of SynGAP1.Brief Bioinform. 2024 Sep 23;25(6):bbae458. doi: 10.1093/bib/bbae458. Brief Bioinform. 2024. PMID: 39311700 Free PMC article.
-
Induced Coma, Death, and Organ Transplantation: A Physiologic, Genetic, and Theological Perspective.Int J Mol Sci. 2023 Mar 17;24(6):5744. doi: 10.3390/ijms24065744. Int J Mol Sci. 2023. PMID: 36982814 Free PMC article. Review.
-
New insights into RAS in head and neck cancer.Biochim Biophys Acta Rev Cancer. 2023 Nov;1878(6):188963. doi: 10.1016/j.bbcan.2023.188963. Epub 2023 Aug 22. Biochim Biophys Acta Rev Cancer. 2023. PMID: 37619805 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

