The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean
- PMID: 33096644
- PMCID: PMC7589023
- DOI: 10.3390/ijms21207778
The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean
Abstract
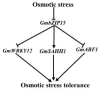
Soybean (Glycine max), as an important oilseed crop, is constantly threatened by abiotic stress, including that caused by salinity and drought. bZIP transcription factors (TFs) are one of the largest TF families and have been shown to be associated with various environmental-stress tolerances among species; however, their function in abiotic-stress response in soybean remains poorly understood. Here, we characterized the roles of soybean transcription factor GmbZIP15 in response to abiotic stresses. The transcript level of GmbZIP15 was suppressed under salt- and drought-stress conditions. Overexpression of GmbZIP15 in soybean resulted in hypersensitivity to abiotic stress compared with wild-type (WT) plants, which was associated with lower transcript levels of stress-responsive genes involved in both abscisic acid (ABA)-dependent and ABA-independent pathways, defective stomatal aperture regulation, and reduced antioxidant enzyme activities. Furthermore, plants expressing a functional repressor form of GmbZIP15 exhibited drought-stress resistance similar to WT. RNA-seq and qRT-PCR analyses revealed that GmbZIP15 positively regulates GmSAHH1 expression and negatively regulates GmWRKY12 and GmABF1 expression in response to abiotic stress. Overall, these data indicate that GmbZIP15 functions as a negative regulator in response to salt and drought stresses.
Keywords: GmbZIP15; RNA-seq; drought stress; salt stress; soybean; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

