Circular Intensity Differential Scattering for Label-Free Chromatin Characterization: A Review for Optical Microscopy
- PMID: 33096877
- PMCID: PMC7588990
- DOI: 10.3390/polym12102428
Circular Intensity Differential Scattering for Label-Free Chromatin Characterization: A Review for Optical Microscopy
Abstract
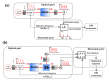
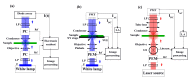
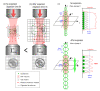
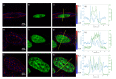
Circular Intensity Differential Scattering (CIDS) provides a differential measurement of the circular right and left polarized light and has been proven to be a gold standard label-free technique to study the molecular conformation of complex biopolymers, such as chromatin. In early works, it has been shown that the scattering component of the CIDS signal gives information from the long-range chiral organization on a scale down to 1/10th-1/20th of the excitation wavelength, leading to information related to the structure and orientation of biopolymers in situ at the nanoscale. In this paper, we review the typical methods and technologies employed for measuring this signal coming from complex macro-molecules ordering. Additionally, we include a general description of the experimental architectures employed for spectroscopic CIDS measurements, angular or spectral, and of the most recent advances in the field of optical imaging microscopy, allowing a visualization of the chromatin organization in situ.
Keywords: DNA organization; biopolymers; microscopy; polarization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Combined approach using circular intensity differential scattering microscopy under phasor map data analysis.Appl Opt. 2021 Feb 20;60(6):1558-1565. doi: 10.1364/AO.417677. Appl Opt. 2021. PMID: 33690489
-
Circular intensity differential scattering and chromatin-DNA structure. A combined theoretical approach.Cell Biophys. 1987 Feb;10(1):45-60. doi: 10.1007/BF02797073. Cell Biophys. 1987. PMID: 2440578
-
Differential scattering of circularly polarized light as a unique probe of polynucleosome superstructures. A simulation by multiple scattering of dipoles.Cell Biophys. 1983 Sep;5(3):163-87. doi: 10.1007/BF02788611. Cell Biophys. 1983. PMID: 6199111
-
Circular differential scattering can be an important part of the circular dichroism of macromolecules.Proc Natl Acad Sci U S A. 1983 Jun;80(12):3568-72. doi: 10.1073/pnas.80.12.3568. Proc Natl Acad Sci U S A. 1983. PMID: 6574499 Free PMC article.
-
Recent Trends in Chiroptical Spectroscopy: Theory and Applications of Vibrational Circular Dichroism and Raman Optical Activity.Chempluschem. 2020 Mar;85(3):561-575. doi: 10.1002/cplu.202000014. Epub 2020 Mar 18. Chempluschem. 2020. PMID: 32187832 Review.
Cited by
-
Computational Modeling of Chromatin Fiber to Characterize Its Organization Using Angle-Resolved Scattering of Circularly Polarized Light.Polymers (Basel). 2021 Oct 5;13(19):3422. doi: 10.3390/polym13193422. Polymers (Basel). 2021. PMID: 34641237 Free PMC article.
-
Multiparametric Remote Investigation in the near-IR through Optical Fiber for In Situ Measurements.Sensors (Basel). 2023 Mar 7;23(6):2911. doi: 10.3390/s23062911. Sensors (Basel). 2023. PMID: 36991622 Free PMC article.
-
Orientation-independent-DIC imaging reveals that a transient rise in depletion attraction contributes to mitotic chromosome condensation.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2403153121. doi: 10.1073/pnas.2403153121. Epub 2024 Aug 27. Proc Natl Acad Sci U S A. 2024. PMID: 39190347 Free PMC article.
-
Phasor map analysis to investigate Hutchinson-Gilford progeria cell under polarization-resolved optical scanning microscopy.Sci Rep. 2022 Jan 31;12(1):1679. doi: 10.1038/s41598-022-05755-1. Sci Rep. 2022. PMID: 35102338 Free PMC article.
References
-
- Huard S. Polarization of Light. 1st ed. John Wiley & Sons Inc.; New York, NY, USA: 1997. Polarization of Light.
Publication types
LinkOut - more resources
Full Text Sources

