Advanced Methods for Studying Structure and Interactions of Macrolide Antibiotics
- PMID: 33096889
- PMCID: PMC7589898
- DOI: 10.3390/ijms21207799
Advanced Methods for Studying Structure and Interactions of Macrolide Antibiotics
Abstract
Macrolide antibiotics are macrocyclic compounds that are clinically used and prescribed for the treatment of upper and lower respiratory tract infections. They inhibit the synthesis of bacterial proteins by reversible binding to the 23S rRNA at or near the peptidyl transferase center. However, their excellent antibacterial profile was largely compromised by the emergence of bacterial resistance. Today, fighting resistance to antibiotics is one of the greatest challenges in medicinal chemistry. Considering various physicochemical properties of macrolides, understanding their structure and interactions with macromolecular targets is crucial for the design of new antibiotics efficient against resistant pathogens. The solid-state structures of some macrolide-ribosome complexes have recently been solved, throwing new light on the macrolide binding mechanisms. On the other hand, a combination of NMR spectroscopy and molecular modeling calculations can be applied to study free and bound conformations in solution. In this article, a description of advanced physicochemical methods for elucidating the structure and interactions of macrolide antibiotics in solid state and solution will be provided, and their principal advantages and drawbacks will be discussed.
Keywords: NMR spectroscopy; X-ray crystallography; biochemical and fluorescence methods; biomolecular targets; cryo-electron microscopy; macrolide antibiotics; macrolide interactions; molecular dynamics simulations; structure characterization.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

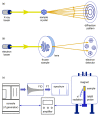
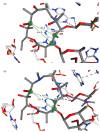


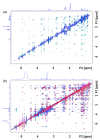
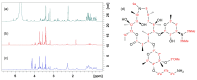

Similar articles
-
Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains.Life Sci Alliance. 2025 Jun 9;8(8):e202503325. doi: 10.26508/lsa.202503325. Print 2025 Aug. Life Sci Alliance. 2025. PMID: 40490363 Free PMC article.
-
Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action.Ann N Y Acad Sci. 2011 Dec;1241:33-47. doi: 10.1111/j.1749-6632.2011.06315.x. Ann N Y Acad Sci. 2011. PMID: 22191525 Review.
-
Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA.J Bacteriol. 2001 Dec;183(23):6898-907. doi: 10.1128/JB.183.23.6898-6907.2001. J Bacteriol. 2001. PMID: 11698379 Free PMC article.
-
Structural consideration of macrolide antibiotics in relation to the ribosomal interaction and drug design.Curr Top Med Chem. 2003;3(9):991-9. doi: 10.2174/1568026033452177. Curr Top Med Chem. 2003. PMID: 12678833 Review.
-
Context-specific action of macrolide antibiotics on the eukaryotic ribosome.Nat Commun. 2021 May 14;12(1):2803. doi: 10.1038/s41467-021-23068-1. Nat Commun. 2021. PMID: 33990576 Free PMC article.
Cited by
-
SPR741, Double- or Triple-Combined With Erythromycin and Clarithromycin, Combats Drug-Resistant Klebsiella pneumoniae, Its Biofilms, and Persister Cells.Front Cell Infect Microbiol. 2022 Mar 18;12:858606. doi: 10.3389/fcimb.2022.858606. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35372124 Free PMC article.
-
Synergistic Effects of Azithromycin and STING Agonist Promote IFN-I Production by Enhancing the Activation of STING-TBK1 Signaling.J Exp Pharmacol. 2023 Nov 1;15:407-421. doi: 10.2147/JEP.S433181. eCollection 2023. J Exp Pharmacol. 2023. PMID: 37933302 Free PMC article.
-
The Crisis of Macrolide Resistance in Pneumococci in Latin America.Am J Trop Med Hyg. 2024 Jul 30;111(4):756-764. doi: 10.4269/ajtmh.23-0913. Print 2024 Oct 2. Am J Trop Med Hyg. 2024. PMID: 39084209 Free PMC article. Review.
-
The Physical Chemistry and Chemical Physics (PCCP) Section of the International Journal of Molecular Sciences in Its Publications: The First 300 Thematic Articles in the First 3 Years.Int J Mol Sci. 2021 Dec 27;23(1):241. doi: 10.3390/ijms23010241. Int J Mol Sci. 2021. PMID: 35008667 Free PMC article.
-
Interactions of Aminopropyl-Azithromycin Derivatives, Precursors in the Synthesis of Bioactive Macrozones, with E. coli Ribosome: NMR and Docking Studies.Materials (Basel). 2021 Sep 25;14(19):5561. doi: 10.3390/ma14195561. Materials (Basel). 2021. PMID: 34639957 Free PMC article.
References
-
- Arsić B., Barber J., Novak P. The macrolide antibiotics and their semi-synthetic derivatives. In: Arsić B., editor. Macrolides: Properties, Synthesis and Applications. Walter de Gruyter GmbH; Berlin, Germany: Boston, MA, USA: 2018. pp. 1–30. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

