How HIV-1 Integrase Associates with Human Mitochondrial Lysyl-tRNA Synthetase
- PMID: 33096929
- PMCID: PMC7589778
- DOI: 10.3390/v12101202
How HIV-1 Integrase Associates with Human Mitochondrial Lysyl-tRNA Synthetase
Abstract
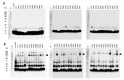
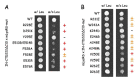
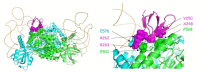
Replication of human immunodeficiency virus type 1 (HIV-1) requires the packaging of tRNALys,3 from the host cell into the new viral particles. The GagPol viral polyprotein precursor associates with mitochondrial lysyl-tRNA synthetase (mLysRS) in a complex with tRNALys, an essential step to initiate reverse transcription in the virions. The C-terminal integrase moiety of GagPol is essential for its association with mLysRS. We show that integrases from HIV-1 and HIV-2 bind mLysRS with the same efficiency. In this work, we have undertaken to probe the three-dimensional (3D) architecture of the complex of integrase with mLysRS. We first established that the C-terminal domain (CTD) of integrase is the major interacting domain with mLysRS. Using the pBpa-photo crosslinking approach, inter-protein cross-links were observed involving amino acid residues located at the surface of the catalytic domain of mLysRS and of the CTD of integrase. In parallel, using molecular docking simulation, a single structural model of complex was found to outscore other alternative conformations. Consistent with crosslinking experiments, this structural model was further probed experimentally. Five compensatory mutations in the two partners were successfully designed which supports the validity of the model. The complex highlights that binding of integrase could stabilize the tRNALys:mLysRS interaction.
Keywords: 3D model; HIV-1; integrase; mitochondrial lysyl-tRNA synthetase; tRNA packaging complex.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures







References
-
- Abbink T.E., Berkhout B. HIV-1 reverse transcription: Close encounters between the viral genome and a cellular tRNA. Adv. Pharmacol. 2007;55:99–135. - PubMed
-
- Gabor J., Cen S., Javanbakht H., Niu M.J., Kleiman L. Effect of altering the tRNA3Lys concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 2002;76:9096–9102. doi: 10.1128/JVI.76.18.9096-9102.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

