Fragment Exchange Plasmid Tools for CRISPR/Cas9-Mediated Gene Integration and Protease Production in Bacillus subtilis
- PMID: 33097498
- PMCID: PMC7755240
- DOI: 10.1128/AEM.02090-20
Fragment Exchange Plasmid Tools for CRISPR/Cas9-Mediated Gene Integration and Protease Production in Bacillus subtilis
Abstract
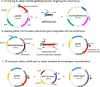
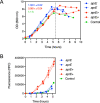
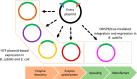
Since its discovery as part of the bacterial adaptative immune system, CRISPR/Cas has emerged as the most promising tool for targeted genome editing over the past few years. Various tools for genome editing in Bacillus subtilis have recently been developed, expanding and simplifying its potential development as an industrial species. A collection of vectors compatible with high-throughput (HTP) fragment exchange (FX) cloning for heterologous expression in Escherichia coli and Bacillus was previously developed. This vector catalogue was through this work supplemented with editing plasmids for genome engineering in Bacillus by adapting two CRISPR/Cas plasmids to the cloning technology. The customized tools allow versatile editing at any chosen genomic position (single-plasmid strategy) or at a fixed genomic locus (double-plasmid strategy). The single-plasmid strategy was validated by deleting the spoIIAC gene, which has an essential role in sporulation. Using the double-plasmid strategy, we demonstrate the quick transition from plasmid-based subtilisin expression to the stable integration of the gene into the amyE locus of a seven-protease-deficient KO7 strain. The newly engineered B. subtilis strain allowed the successful production of a functional enzyme. The customized tools provide improvements to the cloning procedure, should be useful for versatile genomic engineering, and contribute to a cloning platform for a quick transition from HTP enzyme expression to production through the fermentation of industrially relevant B. subtilis and related strains.IMPORTANCE We complemented a cloning platform with new editing plasmids that allow a quick transition from high-throughput cloning and the expression of new enzymes to the stable integration of genes for the production of enzymes through B. subtilis fermentation. We present two systems for the effective assembly cloning of any genome-editing cassette that shortens the engineering procedure to obtain the final editing constructs. The utility of the customized tools is demonstrated by disrupting Bacillus' capacity to sporulate and by introducing the stable expression of subtilisin. The tools should be useful to engineer B. subtilis strains by a variety of recombination events to ultimately improve the application range of this industry-relevant host.
Keywords: Bacillus subtilis; CRISPR; FX cloning; genome editing; microbial fermentation; protease; subtilisin.
Copyright © 2020 García-Moyano et al.
Figures


References
-
- Liu L, Liu Y, Shin HD, Chen RR, Wang NS, Li J, Du G, Chen J. 2013. Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl Microbiol Biotechnol 97:6113–6127. doi:10.1007/s00253-013-4960-4. - DOI - PubMed
-
- Nguyen TT, Quyen TD, Le HT. 2013. Cloning and enhancing production of a detergent- and organic-solvent-resistant nattokinase from Bacillus subtilis VTCC-DVN-12-01 by using an eight-protease-gene-deficient Bacillus subtilis WB800. Microb Cell Fact 12:79. doi:10.1186/1475-2859-12-79. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

