Carbamate C-N Hydrolase Gene ameH Responsible for the Detoxification Step of Methomyl Degradation in Aminobacter aminovorans Strain MDW-2
- PMID: 33097501
- PMCID: PMC7755249
- DOI: 10.1128/AEM.02005-20
Carbamate C-N Hydrolase Gene ameH Responsible for the Detoxification Step of Methomyl Degradation in Aminobacter aminovorans Strain MDW-2
Abstract
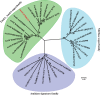
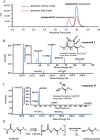
Methomyl {bis[1-methylthioacetaldehyde-O-(N-methylcarbamoyl)oximino]sulfide} is a highly toxic oxime carbamate insecticide. Several methomyl-degrading microorganisms have been reported so far, but the role of specific enzymes and genes in this process is still unexplored. In this study, a protein annotated as a carbamate C-N hydrolase was identified in the methomyl-degrading strain Aminobacter aminovorans MDW-2, and the encoding gene was termed ameH A comparative analysis between the mass fingerprints of AmeH and deduced proteins of the strain MDW-2 genome revealed AmeH to be a key enzyme of the detoxification step of methomyl degradation. The results also demonstrated that AmeH was a functional homodimer with a subunit molecular mass of approximately 34 kDa and shared the highest identity (27%) with the putative formamidase from Schizosaccharomyces pombe ATCC 24843. AmeH displayed maximal enzymatic activity at 50°C and pH 8.5. Km and kcat of AmeH for methomyl were 87.5 μM and 345.2 s-1, respectively, and catalytic efficiency (kcat/Km ) was 3.9 μM-1 s-1 Phylogenetic analysis revealed AmeH to be a member of the FmdA_AmdA superfamily. Additionally, five key amino acid residues (162, 164, 191, 193, and 207) of AmeH were identified by amino acid variations.IMPORTANCE Based on the structural characteristic, carbamate insecticides can be classified into oxime carbamates (methomyl, aldicarb, oxamyl, etc.) and N-methyl carbamates (carbaryl, carbofuran, isoprocarb, etc.). So far, research on the degradation of carbamate pesticides has mainly focused on the detoxification step and hydrolysis of their carbamate bond. Several genes, such as cehA, mcbA, cahA, and mcd, and their encoding enzymes have also been reported to be involved in the detoxification step. However, none of these enzymes can hydrolyze methomyl. In this study, a carbamate C-N hydrolase gene, ameH, responsible for the detoxification step of methomyl in strain MDW-2 was cloned and the key amino acid sites of AmeH were investigated. These findings provide insight into the microbial degradation mechanism of methomyl.
Keywords: Aminobacter aminovorans MDW-2; biodegradation; carbamate C-N hydrolase; methomyl.
Copyright © 2020 American Society for Microbiology.
Figures
Similar articles
-
Degradation of methomyl by the combination of Aminobacter sp. MDW-2 and Afipia sp. MDW-3.Lett Appl Microbiol. 2017 Apr;64(4):289-296. doi: 10.1111/lam.12715. Epub 2017 Feb 27. Lett Appl Microbiol. 2017. PMID: 28083911
-
Identification of the key amino acid sites of the carbofuran hydrolase CehA from a newly isolated carbofuran-degrading strain Sphingbium sp. CFD-1.Ecotoxicol Environ Saf. 2020 Feb;189:109938. doi: 10.1016/j.ecoenv.2019.109938. Epub 2019 Nov 20. Ecotoxicol Environ Saf. 2020. PMID: 31759739
-
Isolation of Oxamyl-degrading Bacteria and Identification of cehA as a Novel Oxamyl Hydrolase Gene.Front Microbiol. 2016 Apr 29;7:616. doi: 10.3389/fmicb.2016.00616. eCollection 2016. Front Microbiol. 2016. PMID: 27199945 Free PMC article.
-
Insights into the microbial degradation and biochemical mechanisms of carbamates.Chemosphere. 2021 Sep;279:130500. doi: 10.1016/j.chemosphere.2021.130500. Epub 2021 Apr 7. Chemosphere. 2021. PMID: 33892453 Review.
-
Current Approaches to and Future Perspectives on Methomyl Degradation in Contaminated Soil/Water Environments.Molecules. 2020 Feb 8;25(3):738. doi: 10.3390/molecules25030738. Molecules. 2020. PMID: 32046287 Free PMC article. Review.
Cited by
-
The Novel Amidase PcnH Initiates the Degradation of Phenazine-1-Carboxamide in Sphingomonas histidinilytica DS-9.Appl Environ Microbiol. 2022 Jun 14;88(11):e0054322. doi: 10.1128/aem.00543-22. Epub 2022 May 17. Appl Environ Microbiol. 2022. PMID: 35579476 Free PMC article.
-
Microbial adaptation and impact into the pesticide's degradation.Arch Microbiol. 2022 Apr 28;204(5):288. doi: 10.1007/s00203-022-02899-6. Arch Microbiol. 2022. PMID: 35482163 Review.
-
Conserved Metabolic and Evolutionary Themes in Microbial Degradation of Carbamate Pesticides.Front Microbiol. 2021 Jul 7;12:648868. doi: 10.3389/fmicb.2021.648868. eCollection 2021. Front Microbiol. 2021. PMID: 34305823 Free PMC article. Review.
References
-
- El-Fakharany II, Massoud AH, Derbalah AS, Allah MSS. 2011. Toxicological effects of methomyl and remediation technologies of its residues in an aquatic system. J Environ Chem Ecotoxicol 3:332–339.
-
- Chen CS, Wu TW, Wang HL, Wu SH, Tien CJ. 2015. The ability of immobilized bacterial consortia and strains from river biofilms to degrade the carbamate pesticide methomyl. Int J Environ Sci Technol 12:2857–2866. doi:10.1007/s13762-014-0675-z. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

