Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer's disease
- PMID: 33097708
- PMCID: PMC7584603
- DOI: 10.1038/s41467-020-19227-5
Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer's disease
Erratum in
-
Author Correction: Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer's disease.Nat Commun. 2023 Mar 2;14(1):1194. doi: 10.1038/s41467-023-36930-1. Nat Commun. 2023. PMID: 36864071 Free PMC article. No abstract available.
Abstract
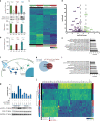
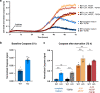
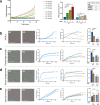
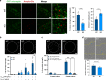
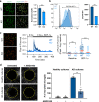
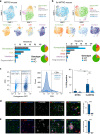
The discovery of TREM2 as a myeloid-specific Alzheimer's disease (AD) risk gene has accelerated research into the role of microglia in AD. While TREM2 mouse models have provided critical insight, the normal and disease-associated functions of TREM2 in human microglia remain unclear. To examine this question, we profile microglia differentiated from isogenic, CRISPR-modified TREM2-knockout induced pluripotent stem cell (iPSC) lines. By combining transcriptomic and functional analyses with a chimeric AD mouse model, we find that TREM2 deletion reduces microglial survival, impairs phagocytosis of key substrates including APOE, and inhibits SDF-1α/CXCR4-mediated chemotaxis, culminating in an impaired response to beta-amyloid plaques in vivo. Single-cell sequencing of xenotransplanted human microglia further highlights a loss of disease-associated microglial (DAM) responses in human TREM2 knockout microglia that we validate by flow cytometry and immunohistochemistry. Taken together, these studies reveal both conserved and novel aspects of human TREM2 biology that likely play critical roles in the development and progression of AD.
Conflict of interest statement
M.B.J. is a co-inventor of patent WO/2018/160496 related to differentiation of human pluripotent stem cells into microglia. All other authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI121945/AI/NIAID NIH HHS/United States
- RF1 AG048099/AG/NIA NIH HHS/United States
- S10 OD021718/OD/NIH HHS/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- T32 AG000096/AG/NIA NIH HHS/United States
- R01 AG061895/AG/NIA NIH HHS/United States
- S10 OD010794/OD/NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- T32 NS082174/NS/NINDS NIH HHS/United States
- R01 AG056303/AG/NIA NIH HHS/United States
- S10 RR025496/RR/NCRR NIH HHS/United States
- R15 AG059236/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- R01 NS014609/NS/NINDS NIH HHS/United States
- F30 AG060704/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

