Single molecule, near full-length genome sequencing of dengue virus
- PMID: 33097792
- PMCID: PMC7584602
- DOI: 10.1038/s41598-020-75374-1
Single molecule, near full-length genome sequencing of dengue virus
Abstract
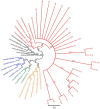
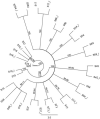
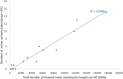
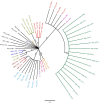
Current methods for dengue virus (DENV) genome amplification, amplify parts of the genome in at least 5 overlapping segments and then combine the output to characterize a full genome. This process is laborious, costly and requires at least 10 primers per serotype, thus increasing the likelihood of PCR bias. We introduce an assay to amplify near full-length dengue virus genomes as intact molecules, sequence these amplicons with third generation "nanopore" technology without fragmenting and use the sequence data to differentiate within-host viral variants with a bioinformatics tool (Nano-Q). The new assay successfully generated near full-length amplicons from DENV serotypes 1, 2 and 3 samples which were sequenced with nanopore technology. Consensus DENV sequences generated by nanopore sequencing had over 99.5% pairwise sequence similarity to Illumina generated counterparts provided the coverage was > 100 with both platforms. Maximum likelihood phylogenetic trees generated from nanopore consensus sequences were able to reproduce the exact trees made from Illumina sequencing with a conservative 99% bootstrapping threshold (after 1000 replicates and 10% burn-in). Pairwise genetic distances of within host variants identified from the Nano-Q tool were less than that of between host variants, thus enabling the phylogenetic segregation of variants from the same host.
Conflict of interest statement
Martin A. Smith: I have read the journal's policy and the authors of this manuscript have the following competing interests: I have received travel support to speak at conferences from Oxford Nanopore Technologies. All other authors have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

