Lung gene expression signatures suggest pathogenic links and molecular markers for pulmonary tuberculosis, adenocarcinoma and sarcoidosis
- PMID: 33097805
- PMCID: PMC7584606
- DOI: 10.1038/s42003-020-01318-0
Lung gene expression signatures suggest pathogenic links and molecular markers for pulmonary tuberculosis, adenocarcinoma and sarcoidosis
Abstract
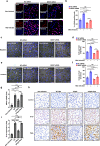
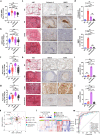
Previous reports have suggested a link between pulmonary tuberculosis (TB), which is caused by Mycobacterium tuberculosis (Mtb), and the development of lung adenocarcinoma (LUAD) and sarcoidosis. Furthermore, these lung diseases share certain clinical similarities that can challenge differential diagnosis in some cases. Here, through comparison of lung transcriptome-derived molecular signatures of TB, LUAD and sarcoidosis patients, we identify certain shared disease-related expression patterns. We also demonstrate that MKI67, an over-expressed gene shared by TB and LUAD, is a key mediator in Mtb-promoted tumor cell proliferation, migration, and invasion. Moreover, we reveal a distinct ossification-related TB lung signature, which may be associated with the activation of the BMP/SMAD/RUNX2 pathway in Mtb-infected macrophages that can restrain mycobacterial survival and promote osteogenic differentiation of mesenchymal stem cells. Taken together, these findings provide novel pathogenic links and potential molecular markers for better understanding and differential diagnosis of pulmonary TB, LUAD and sarcoidosis.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- World Health Organization. Global Tuberculosis Report 2019. (WHO, Geneva, 2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

