Monogenic variants in dystonia: an exome-wide sequencing study
- PMID: 33098801
- PMCID: PMC8246240
- DOI: 10.1016/S1474-4422(20)30312-4
Monogenic variants in dystonia: an exome-wide sequencing study
Abstract
Background: Dystonia is a clinically and genetically heterogeneous condition that occurs in isolation (isolated dystonia), in combination with other movement disorders (combined dystonia), or in the context of multisymptomatic phenotypes (isolated or combined dystonia with other neurological involvement). However, our understanding of its aetiology is still incomplete. We aimed to elucidate the monogenic causes for the major clinical categories of dystonia.
Methods: For this exome-wide sequencing study, study participants were identified at 33 movement-disorder and neuropaediatric specialty centres in Austria, Czech Republic, France, Germany, Poland, Slovakia, and Switzerland. Each individual with dystonia was diagnosed in accordance with the dystonia consensus definition. Index cases were eligible for this study if they had no previous genetic diagnosis and no indication of an acquired cause of their illness. The second criterion was not applied to a subset of participants with a working clinical diagnosis of dystonic cerebral palsy. Genomic DNA was extracted from blood of participants and whole-exome sequenced. To find causative variants in known disorder-associated genes, all variants were filtered, and unreported variants were classified according to American College of Medical Genetics and Genomics guidelines. All considered variants were reviewed in expert round-table sessions to validate their clinical significance. Variants that survived filtering and interpretation procedures were defined as diagnostic variants. In the cases that went undiagnosed, candidate dystonia-causing genes were prioritised in a stepwise workflow.
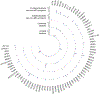
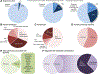
Findings: We sequenced the exomes of 764 individuals with dystonia and 346 healthy parents who were recruited between June 1, 2015, and July 31, 2019. We identified causative or probable causative variants in 135 (19%) of 728 families, involving 78 distinct monogenic disorders. We observed a larger proportion of individuals with diagnostic variants in those with dystonia (either isolated or combined) with coexisting non-movement disorder-related neurological symptoms (100 [45%] of 222; excepting cases with evidence of perinatal brain injury) than in those with combined (19 [19%] of 98) or isolated (16 [4%] of 388) dystonia. Across all categories of dystonia, 104 (65%) of the 160 detected variants affected genes which are associated with neurodevelopmental disorders. We found diagnostic variants in 11 genes not previously linked to dystonia, and propose a predictive clinical score that could guide the implementation of exome sequencing in routine diagnostics. In cases without perinatal sentinel events, genomic alterations contributed substantively to the diagnosis of dystonic cerebral palsy. In 15 families, we delineated 12 candidate genes. These include IMPDH2, encoding a key purine biosynthetic enzyme, for which robust evidence existed for its involvement in a neurodevelopmental disorder with dystonia. We identified six variants in IMPDH2, collected from four independent cohorts, that were predicted to be deleterious de-novo variants and expected to result in deregulation of purine metabolism.
Interpretation: In this study, we have determined the role of monogenic variants across the range of dystonic disorders, providing guidance for the introduction of personalised care strategies and fostering follow-up pathophysiological explorations.
Funding: Else Kröner-Fresenius-Stiftung, Technische Universität München, Helmholtz Zentrum München, Medizinische Universität Innsbruck, Charles University in Prague, Czech Ministry of Education, the Slovak Grant and Development Agency, the Slovak Research and Grant Agency.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests
R.J. received honoraria and consultancies from Medtronic, Cardion, Ipsen and NeuroPa centrum. A.J. reports personal fees and non-financial support from Pharm Allergan GmbH, non-financial support from Boston Scientific GmbH, non-financial support from Ipsen Pharma Gmbh, non-financial support from Merz Pharmaceuticals GmbH, and non-financial support from Bayer Vital GmbH. Y. D. reports personal fees from Zentrum für Humangenetik und Laboratoriumsdiagnostik (MVZ), Martinsried, Germany. Z.G. received payments for lectures from Bayer, Biogen, Boehringer-Ingelheim, MSD, Novartis, Pfizer, Sandoz, Shire, and TEVA. J. Š. reports personal fees from Medtronic. A.T., R.E.P., F.M.Z., and L.B.H. are employees of GeneDx, Inc. P.G.-A. is a principal investigator in the NeuroSeq study and received consulting fees from Spark Therapeutics, Eisai Therapeutics, and NeuExcell Therapeutics. The other authors declare no competing interests.
Figures


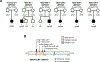
Comment in
-
Dystonia: genetics, phenomenology, and pathophysiology.Lancet Neurol. 2020 Nov;19(11):881-882. doi: 10.1016/S1474-4422(20)30366-5. Lancet Neurol. 2020. PMID: 33098786 No abstract available.
-
Monogenic Causes of Dystonic Syndromes: Common in Dystonic Cerebral Palsy, Rare in Isolated Dystonia.Mov Disord. 2021 Jan;36(1):84. doi: 10.1002/mds.28420. Epub 2020 Dec 7. Mov Disord. 2021. PMID: 33284469 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

