Genomic evidence for recurrent genetic admixture during the domestication of Mediterranean olive trees (Olea europaea L.)
- PMID: 33100219
- PMCID: PMC7586694
- DOI: 10.1186/s12915-020-00881-6
Genomic evidence for recurrent genetic admixture during the domestication of Mediterranean olive trees (Olea europaea L.)
Abstract
Background: Olive tree (Olea europaea L. subsp. europaea, Oleaceae) has been the most emblematic perennial crop for Mediterranean countries since its domestication around 6000 years ago in the Levant. Two taxonomic varieties are currently recognized: cultivated (var. europaea) and wild (var. sylvestris) trees. However, it remains unclear whether olive cultivars derive from a single initial domestication event followed by secondary diversification, or whether cultivated lineages are the result of more than a single, independent primary domestication event. To shed light into the recent evolution and domestication of the olive tree, here we analyze a group of newly sequenced and available genomes using a phylogenomics and population genomics framework.
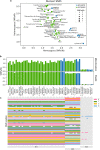
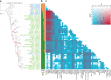
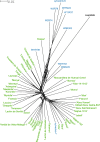
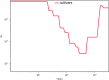
Results: We improved the assembly and annotation of the reference genome, newly sequenced the genomes of twelve individuals: ten var. europaea, one var. sylvestris, and one outgroup taxon (subsp. cuspidata)-and assembled a dataset comprising whole genome data from 46 var. europaea and 10 var. sylvestris. Phylogenomic and population structure analyses support a continuous process of olive tree domestication, involving a major domestication event, followed by recurrent independent genetic admixture events with wild populations across the Mediterranean Basin. Cultivated olives exhibit only slightly lower levels of genetic diversity than wild forms, which can be partially explained by the occurrence of a mild population bottleneck 3000-14,000 years ago during the primary domestication period, followed by recurrent introgression from wild populations. Genes associated with stress response and developmental processes were positively selected in cultivars, but we did not find evidence that genes involved in fruit size or oil content were under positive selection. This suggests that complex selective processes other than directional selection of a few genes are in place.
Conclusions: Altogether, our results suggest that a primary domestication area in the eastern Mediterranean basin was followed by numerous secondary events across most countries of southern Europe and northern Africa, often involving genetic admixture with genetically rich wild populations, particularly from the western Mediterranean Basin.
Keywords: Admixture; Domestication; Genome; Introgression; Olive.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Green PS. A revision of olea L. (oleaceae) Kew Bull. 2002;57:91. doi: 10.2307/4110824. - DOI
-
- Vargas P, Kadereit JW. Molecular fingerprinting evidence (ISSR, Inter-Simple Sequence Repeats) for a wild status of Olea europaea L. (Oleaceae) in the Eurosiberian North of the Iberian Peninsula. Flora. 2001;196:142–152. doi: 10.1016/S0367-2530(17)30029-4. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

