Potentials and Challenges of Genomics for Breeding Cannabis Cultivars
- PMID: 33101342
- PMCID: PMC7546024
- DOI: 10.3389/fpls.2020.573299
Potentials and Challenges of Genomics for Breeding Cannabis Cultivars
Abstract
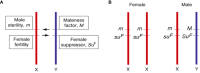
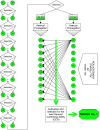
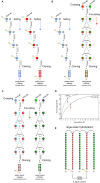
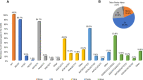
Cannabis (Cannabis sativa L.) is an influential yet controversial agricultural plant with a very long and prominent history of recreational, medicinal, and industrial usages. Given the importance of this species, we deepened some of the main challenges-along with potential solutions-behind the breeding of new cannabis cultivars. One of the main issues that should be fixed before starting new breeding programs is the uncertain taxonomic classification of the two main taxa (e.g., indica and sativa) of the Cannabis genus. We tried therefore to examine this topic from a molecular perspective through the use of DNA barcoding. Our findings seem to support a unique species system (C. sativa) based on two subspecies: C. sativa subsp. sativa and C. sativa subsp. indica. The second key issue in a breeding program is related to the dioecy behavior of this species and to the comprehension of those molecular mechanisms underlying flower development, the main cannabis product. Given the role of MADS box genes in flower identity, we analyzed and reorganized all the genomic and transcriptomic data available for homeotic genes, trying to decipher the applicability of the ABCDE model in Cannabis. Finally, reviewing the limits of the conventional breeding methods traditionally applied for developing new varieties, we proposed a new breeding scheme for the constitution of F1 hybrids, without ignoring the indisputable contribution offered by genomics. In this sense, in parallel, we resumed the main advances in the genomic field of this species and, ascertained the lack of a robust set of SNP markers, provided a discriminant and polymorphic panel of SSR markers as a valuable tool for future marker assisted breeding programs.
Keywords: Cannabis sativa, breeding methods; DNA barcodes; F1 hybrids; MADS box genes; simple sequence repeat markers.
Copyright © 2020 Barcaccia, Palumbo, Scariolo, Vannozzi, Borin and Bona.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

