Integrated Analyses of Mouse Stem Cell Transcriptomes Provide Clues for Stem Cell Maintenance and Transdifferentiation
- PMID: 33101382
- PMCID: PMC7500244
- DOI: 10.3389/fgene.2020.563798
Integrated Analyses of Mouse Stem Cell Transcriptomes Provide Clues for Stem Cell Maintenance and Transdifferentiation
Abstract
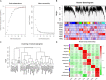
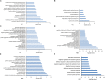
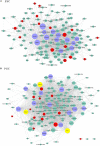
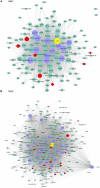
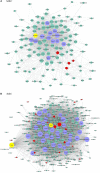
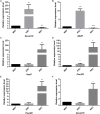
In vivo cell fate reprogramming has emerged as a new method for understanding cell plasticity and as potential treatment for tissue regeneration. Highly efficient and precise reprogramming requires fully understanding of the transcriptomes which function within different cell types. Here, we adopt weighted gene co-expression network analysis (WGCNA) to explore the molecular mechanisms of self-renewal in several well-known stem cell types, including embryonic stem cells (ESC), primordial germ cells (PGC), spermatogonia stem cells (SSC), neural stem cells (NSC), mesenchymal stem cells (MSC), and hematopoietic stem cells (HSC). We identified 37 core genes that were up-regulated in all of the stem cell types examined, as well as stem cell correlated gene co-expression networks. The validation of the co-expression genes revealed a continued protein-protein interaction network that included 823 nodes and 3113 edges. Based on the topology, we identified six densely connected regions within the continued protein-protein interaction network. The SSC specific genes Itgam, Cxcr6, and Agtr2 bridged four densely connected regions that consisted primarily of HSC-, NSC-, and MSC-correlated genes. The expression levels of identified stem cell related transcription factors were confirmed consistent with bioinformatics prediction in ESCs and NSCs by qPCR. Exploring the mechanisms underlying adult stem cell self-renewal will aid in the understanding of stem cell pool maintenance and will promote more accurate and efficient strategies for tissue regeneration and repair.
Keywords: co-expression; network; stem cell; transcriptomes; transdifferentiation.
Copyright © 2020 Wang, Li, Hou, Song, Xie and Shen.
Figures




Similar articles
-
Construction and analysis of a protein-protein interaction network related to self-renewal of mouse spermatogonial stem cells.Mol Biosyst. 2015 Mar;11(3):835-43. doi: 10.1039/c4mb00579a. Epub 2015 Jan 5. Mol Biosyst. 2015. PMID: 25566695
-
Bioinformatics and microarray analysis of microRNA expression profiles of murine embryonic stem cells, neural stem cells induced from ESCs and isolated from E8.5 mouse neural tube.Neurol Res. 2010 Jul;32(6):603-13. doi: 10.1179/174313209X455691. Epub 2009 Aug 5. Neurol Res. 2010. PMID: 19660235
-
Expression profile of an operationally-defined neural stem cell clone.Exp Neurol. 2005 Aug;194(2):320-32. doi: 10.1016/j.expneurol.2005.04.018. Exp Neurol. 2005. PMID: 15992799
-
In or out stemness: comparing growth factor signalling in mouse embryonic stem cells and primordial germ cells.Curr Stem Cell Res Ther. 2009 May;4(2):87-97. doi: 10.2174/157488809788167391. Curr Stem Cell Res Ther. 2009. PMID: 19442193 Review.
-
Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network.Cell Mol Life Sci. 2015 May;72(9):1741-57. doi: 10.1007/s00018-015-1833-2. Epub 2015 Jan 17. Cell Mol Life Sci. 2015. PMID: 25595304 Free PMC article. Review.
Cited by
-
S100A6 as a Constituent and Potential Marker of Adult and Cancer Stem Cells.Stem Cell Rev Rep. 2022 Dec;18(8):2699-2708. doi: 10.1007/s12015-022-10403-2. Epub 2022 Jul 7. Stem Cell Rev Rep. 2022. PMID: 35796891 Review.
-
Macrophage-Secreted CSF1 Transmits a Calorie Restriction-Induced Self-Renewal Signal to Mammary Epithelial Stem Cells.Cells. 2022 Sep 19;11(18):2923. doi: 10.3390/cells11182923. Cells. 2022. PMID: 36139499 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

