Genetic Association of ACE2 rs2285666 Polymorphism With COVID-19 Spatial Distribution in India
- PMID: 33101387
- PMCID: PMC7545580
- DOI: 10.3389/fgene.2020.564741
Genetic Association of ACE2 rs2285666 Polymorphism With COVID-19 Spatial Distribution in India
Abstract
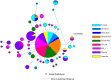
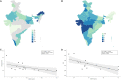
Studies on host-pathogen interaction have identified human ACE2 as a host cell receptor responsible for mediating infection by coronavirus (COVID-19). Subsequent studies have shown striking difference of allele frequency among Europeans and Asians for a polymorphism rs2285666, present in ACE2. It has been revealed that the alternate allele (TT-plus strand or AA-minus strand) of rs2285666 elevate the expression level of this gene upto 50%, hence may play a significant role in SARS-CoV-2 susceptibility. Therefore, we have first looked the phylogenetic structure of rs2285666 derived haplotypes in worldwide populations and compared the spatial frequency of this particular allele with respect to the COVID-19 infection as well as case-fatality rate in India. For the first time, we ascertained a significant positive correlation for alternate allele (T or A) of rs2285666, with the lower infection as well as case-fatality rate among Indian populations. We trust that this information will be useful to understand the role of ACE2 in COVID-19 susceptibility.
Keywords: ACE2; COVID-19; India; SARS-CoV-2; coronavirus; rs2285666.
Copyright © 2020 Srivastava, Bandopadhyay, Das, Pandey, Singh, Khanam, Srivastava, Singh, Dubey, Pathak, Gupta, Rai, Sultana and Chaubey.
Figures
References
-
- Benesty J., Chen J., Huang Y., Cohen I. (2009). “Pearson correlation coefficient,” in Noise Reduction in Speech Processing, Springer Topics in Signal Processing, vol. 2, Berlin, Heidelberg: Springer: 10.1007/978-3-642-00296-0_5 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

