Enrichment and identification of differentially expressed genes in hepatocellular carcinoma stem-like cells
- PMID: 33101493
- PMCID: PMC7577079
- DOI: 10.3892/ol.2020.12162
Enrichment and identification of differentially expressed genes in hepatocellular carcinoma stem-like cells
Abstract
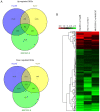
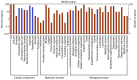
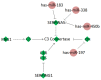
Cancer stem cells are considered to be tumor-initiating cells. To explain the initiation or progression of hepatocellular carcinoma (HCC), we previously established a culture system that may enrich hepatic cancer stem-like cells (HCSCs). However, the regulatory mechanisms by which HCSCs acquire stem cell properties remain unclear. In the present study, three pairs of HCSCs and case-matched human HCC cells were analyzed by high-throughput screening, and novel biomarkers and pathways for the regulation of HCSCs were identified. The results led to the identification and stratification of 406 differentially expressed genes (DEGs), among which 73 GO terms were found to be significantly associated with DEGs in HCSCs, and only complement and coagulation cascade pathways were identified during the development of HCSCs. By combining the results of the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses, it was revealed that 7 genes were downregulated in the complement and coagulation cascade pathways, and 7 miRNAs were predicted to target several downregulated genes involved in these pathways. The results may contribute toward hepatic cancer stem cell studies and novel drug research for HCC treatment.
Keywords: Hepatocellular carcinoma; cancer stem-like cells; differentially expressed genes; high-throughput screening; signaling pathway.
Copyright © 2020, Spandidos Publications.
Figures

Similar articles
-
Long non-coding RNAs and genes contributing to the generation of cancer stem cells in hepatocellular carcinoma identified by RNA sequencing analysis.Oncol Rep. 2016 Nov;36(5):2619-2624. doi: 10.3892/or.2016.5120. Epub 2016 Sep 22. Oncol Rep. 2016. PMID: 27667473
-
Integrated analysis of miRNA, gene, and pathway regulatory networks in hepatic cancer stem cells.J Transl Med. 2015 Aug 11;13:259. doi: 10.1186/s12967-015-0609-7. J Transl Med. 2015. PMID: 26259570 Free PMC article.
-
The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data.Med Oncol. 2017 Jun;34(6):101. doi: 10.1007/s12032-017-0963-9. Epub 2017 Apr 21. Med Oncol. 2017. PMID: 28432618 Free PMC article.
-
The Therapeutic Targets of miRNA in Hepatic Cancer Stem Cells.Stem Cells Int. 2016;2016:1065230. doi: 10.1155/2016/1065230. Epub 2016 Mar 28. Stem Cells Int. 2016. PMID: 27118975 Free PMC article. Review.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
Cited by
-
Role of Hepatocyte-Derived Osteopontin in Liver Carcinogenesis.Hepatol Commun. 2022 Apr;6(4):692-709. doi: 10.1002/hep4.1845. Epub 2021 Nov 3. Hepatol Commun. 2022. PMID: 34730871 Free PMC article.
References
LinkOut - more resources
Full Text Sources
