Metabolic plasticity of IDH1 -mutant glioma cell lines is responsible for low sensitivity to glutaminase inhibition
- PMID: 33101674
- PMCID: PMC7579920
- DOI: 10.1186/s40170-020-00229-2
Metabolic plasticity of IDH1 -mutant glioma cell lines is responsible for low sensitivity to glutaminase inhibition
Abstract
Background: Targeting glutamine metabolism in cancer has become an increasingly vibrant area of research. Mutant IDH1 (IDH1 mut ) gliomas are considered good candidates for targeting this pathway because of the contribution of glutamine to their newly acquired function: synthesis of 2-hydroxyglutarate (2HG).
Methods: We have employed a combination of 13C tracers including glutamine and glucose for investigating the metabolism of patient-derived IDH1 mut glioma cell lines through NMR and LC/MS. Additionally, genetic loss-of-function (in vitro and in vivo) approaches were performed to unravel the adaptability of these cell lines to the inhibition of glutaminase activity.
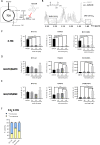
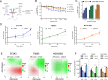
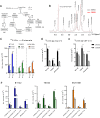
Results: We report the adaptability of IDH1 mut cells' metabolism to the inhibition of glutamine/glutamate pathway. The glutaminase inhibitor CB839 generated a decrease in the production of the downstream metabolites of glutamate, including those involved in the TCA cycle and 2HG. However, this effect on metabolism was not extended to viability; rather, our patient-derived IDH1 mut cell lines display a metabolic plasticity that allows them to overcome glutaminase inhibition.
Conclusions: Major metabolic adaptations involved pathways that can generate glutamate by using alternative substrates from glutamine, such as alanine or aspartate. Indeed, asparagine synthetase was upregulated both in vivo and in vitro revealing a new potential therapeutic target for a combinatory approach with CB839 against IDH1 mut gliomas.
Keywords: 13C tracing; AGI5198; CB839; Gliomas; Glutaminase; IDH1-mutant.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

