Integrative immunogenomic analysis of gastric cancer dictates novel immunological classification and the functional status of tumor-infiltrating cells
- PMID: 33101677
- PMCID: PMC7568758
- DOI: 10.1002/cti2.1194
Integrative immunogenomic analysis of gastric cancer dictates novel immunological classification and the functional status of tumor-infiltrating cells
Abstract
Objectives: A better understanding of antitumor immunity will help predict the prognosis of gastric cancer patients and tailor the appropriate therapies in each patient. Therefore, we propose a novel immunological classification of gastric cancer.
Methods: We performed whole-exome sequencing (WES), RNA-Seq and flow cytometry in 29 gastric cancer patients who received surgery. The TCGA data set of 323 gastric cancer patients and RNA-Seq data of 45 patients who received pembrolizumab (Kim et al. Nat Med 2018; 24: 1449-1458) were also analysed.
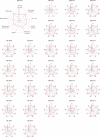
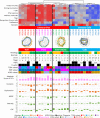
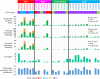
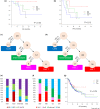
Results: Immunogram analysis of cancer-immunity interaction of gastric cancer revealed immune signatures of four main types, designated Hot1, Hot2, Intermediate and Cold. Immunologically hot tumors displayed a dysfunctional T-cell signature, while cold tumors had an exclusion signature. Ex vivo tumor-infiltrating lymphocyte analysis documented T-cell dysfunction with the expression of checkpoint molecules and impaired cytokine production. The T-cell function was more profoundly damaged in Hot1 than Hot2 tumors. Patients in Hot2 subtypes had better survival in our cohort and TCGA cohort. Although these immunological subtypes overlapped to some degree with the molecular subtypes in the TCGA, intratumoral immune responses cannot be predicted solely based on histological or molecular subtyping of gastric cancer. Molecular and immunological classifications complement each other to predict the responses to anti-PD-1 therapy and have the potential to be a biomarker for the treatment of gastric cancer.
Conclusion: The immunological classification of gastric cancer resulted in four subtypes. Hot tumors were further divided into two subtypes, between which the functional status of T cells was different.
Keywords: RNA‐Seq; T‐cell function; gastric cancer; immunogram; tumor immunity.
© 2020 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
Dr Kakimi reports grants from TAKARA BIO Inc. and grants from MSD, outside the submitted work. In addition, Dr Kakimi has a patent Immunogram pending and the Department of Immunotherapeutics, The University of Tokyo Hospital, is an endowed department by TAKARA BIO Inc. Dr Sato reports personal fees from ONO Pharmaceutical Co., Ltd, Bristol‐Myers Squibb Company, MSD KK and TAIHO Pharmaceutical Co., Ltd, outside the submitted work. Dr Takahashi reports grants and personal fees from Bristol‐Myers Squibb KK, grants and personal fees from ONO Pharmaceutical Co., Ltd, grants and personal fees from MSD, grants and personal fees from AstraZeneca, grants and personal fees from Chugai, and grants and personal fees from BAYER, outside the submitted work. Drs Kakimi and Mineno have a patent Immunogram pending. The other authors have no competing interests to disclose.
Figures


References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. - PubMed
-
- Muro K, Chung HC, Shankaran V et al Pembrolizumab for patients with PD‐L1‐positive advanced gastric cancer (KEYNOTE‐012): a multicentre, open‐label, phase 1b trial. Lancet Oncol 2016; 17: 717–726. - PubMed
-
- Kang YK, Boku N, Satoh T et al Nivolumab in patients with advanced gastric or gastro‐oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO‐4538‐12, ATTRACTION‐2): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet 2017; 390: 2461–2471. - PubMed
-
- Wong SS, Kim KM, Ting JC et al Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole‐genome sequencing. Nat Commun 2014; 5: 5477. - PubMed
LinkOut - more resources
Full Text Sources
