Types I and V Anti-CRISPR Proteins: From Phage Defense to Eukaryotic Synthetic Gene Circuits
- PMID: 33102460
- PMCID: PMC7556299
- DOI: 10.3389/fbioe.2020.575393
Types I and V Anti-CRISPR Proteins: From Phage Defense to Eukaryotic Synthetic Gene Circuits
Abstract
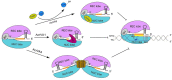
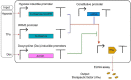
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated proteins), a prokaryotic RNA-mediated adaptive immune system, has been repurposed for gene editing and synthetic gene circuit construction both in bacterial and eukaryotic cells. In the last years, the emergence of the anti-CRISPR proteins (Acrs), which are natural OFF-switches for CRISPR-Cas, has provided a new means to control CRISPR-Cas activity and promoted a further development of CRISPR-Cas-based biotechnological toolkits. In this review, we focus on type I and type V-A anti-CRISPR proteins. We first narrate Acrs discovery and analyze their inhibitory mechanisms from a structural perspective. Then, we describe their applications in gene editing and transcription regulation. Finally, we discuss the potential future usage-and corresponding possible challenges-of these two kinds of anti-CRISPR proteins in eukaryotic synthetic gene circuits.
Keywords: CRISPR; Cas proteins; anti-CRISPR; gene editing; synthetic biology.
Copyright © 2020 Yu and Marchisio.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

