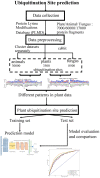
Incorporating Deep Learning With Word Embedding to Identify Plant Ubiquitylation Sites
- PMID: 33102477
- PMCID: PMC7554246
- DOI: 10.3389/fcell.2020.572195
Incorporating Deep Learning With Word Embedding to Identify Plant Ubiquitylation Sites
Abstract
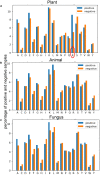
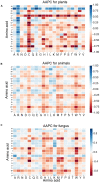
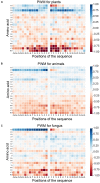
Protein ubiquitylation is an important posttranslational modification (PTM), which is involved in diverse biological processes and plays an essential role in the regulation of physiological mechanisms and diseases. The Protein Lysine Modifications Database (PLMD) has accumulated abundant ubiquitylated proteins with their substrate sites for more than 20 kinds of species. Numerous works have consequently developed a variety of ubiquitylation site prediction tools across all species, mainly relying on the predefined sequence features and machine learning algorithms. However, the difference in ubiquitylated patterns between these species stays unclear. In this work, the sequence-based characterization of ubiquitylated substrate sites has revealed remarkable differences among plants, animals, and fungi. Then an improved word-embedding scheme based on the transfer learning strategy was incorporated with the multilayer convolutional neural network (CNN) for identifying protein ubiquitylation sites. For the prediction of plant ubiquitylation sites, the proposed deep learning scheme could outperform the machine learning-based methods, with the accuracy of 75.6%, precision of 73.3%, recall of 76.7%, F-score of 0.7493, and 0.82 AUC on the independent testing set. Although the ubiquitylated specificity of substrate sites is complicated, this work has demonstrated that the application of the word-embedding method can enable the extraction of informative features and help the identification of ubiquitylated sites. To accelerate the investigation of protein ubiquitylation, the data sets and source code used in this study are freely available at https://github.com/wang-hong-fei/DL-plant-ubsites-prediction.
Keywords: convolutional neural network; deep learning; plant; transfer learning; ubiquitylation; word embedding.
Copyright © 2020 Wang, Wang, Li and Lee.
Figures


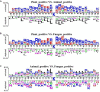
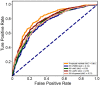
References
-
- Du L., Wang Y., Song G., Lu Z., Wang J. (2018). “Dynamic network embedding: an extended approach for skip-gram based network embedding,” in Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, New York, NY: IJCAI, 2086–2092.
LinkOut - more resources
Full Text Sources

