Base Editing Mediated Generation of Point Mutations Into Human Pluripotent Stem Cells for Modeling Disease
- PMID: 33102492
- PMCID: PMC7546412
- DOI: 10.3389/fcell.2020.590581
Base Editing Mediated Generation of Point Mutations Into Human Pluripotent Stem Cells for Modeling Disease
Abstract
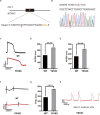
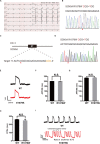
Human pluripotent stem cells (hPSCs) are a powerful platform for disease modeling and drug discovery. However, the introduction of known pathogenic mutations into hPSCs is a time-consuming and labor-intensive process. Base editing is a newly developed technology that enables facile introduction of point mutations into specific loci within the genome of living cells. Here, we design an all-in-one episomal vector that expresses a single guide RNA (sgRNA) with an adenine base editor (ABE) or a cytosine base editor (CBE). Both ABE and CBE can efficiently introduce mutations into cells, A-to-G and C-to-T, respectively. We introduce disease-specific mutations of long QT syndrome into hPSCs to model LQT1, LQT2, and LQT3. Electrophysiological analysis of hPSC-derived cardiomyocytes (hPSC-CMs) using multi-electrode arrays (MEAs) reveals that edited hPSC-CMs display significant increases in duration of the action potential. Finally, we introduce the novel Brugada syndrome-associated mutation into hPSCs, demonstrating that this mutation can cause abnormal electrophysiology. Our study demonstrates that episomal encoded base editors (epi-BEs) can efficiently generate mutation-specific disease hPSC models.
Keywords: Brugada syndrome; IPS; base editing; disease modeling; episomal vector; human pluripotent stem cell; long QT syndrome.
Copyright © 2020 Qi, Wu, Xie, Gao, Li, Pu, Li, Lan and Wang.
Figures



References
-
- Barsheshet A., Goldenberg I., O-Uchi J., Moss A. J., Jons C., Shimizu W., et al. (2012). Mutations in cytoplasmic loops of the KCNQ1 channel and the risk of life-threatening events: implications for mutation-specific response to beta-blocker therapy in type 1 long-QT syndrome. Circulation 125 1988–1996. 10.1161/circulationaha.111.048041 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

