Viral Load and Cell Tropism During Early Latent Equid Herpesvirus 1 Infection Differ Over Time in Lymphoid and Neural Tissue Samples From Experimentally Infected Horses
- PMID: 33102556
- PMCID: PMC7499125
- DOI: 10.3389/fvets.2020.00621
Viral Load and Cell Tropism During Early Latent Equid Herpesvirus 1 Infection Differ Over Time in Lymphoid and Neural Tissue Samples From Experimentally Infected Horses
Abstract
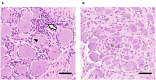
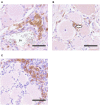
Upper respiratory tract infections with Equid Herpesvirus 1 (EHV-1) typically result in a peripheral blood mononuclear cell-associated viremia, which can lead to vasculopathy in the central nervous system. Primary EHV-1 infection also likely establishes latency in trigeminal ganglia (TG) via retrograde axonal transport and in respiratory tract-associated lymphatic tissue. However, latency establishment and reactivation are poorly understood. To characterize the pathogenesis of EHV-1 latency establishment and maintenance, two separate groups of yearling horses were experimentally infected intranasally with EHV-1, strain Ab4, and euthanized 30 days post infection (dpi), (n = 9) and 70 dpi (n = 6). During necropsy, TG, sympathetic trunk (ST), retropharyngeal and mesenteric lymph nodes (RLn, MesLn) and kidney samples were collected. Viral DNA was detected by quantitative PCR (qPCR) in TG, ST, RLn, and MesLn samples in horses 30 and 70 dpi. The number of positive TG, RLn and MesLn samples was reduced when comparing horses 30 and 70 dpi and the viral copy number in TG and RLn significantly declined from 30 to 70 dpi. EHV-1 late gene glycoprotein B reverse transcriptase PCR and IHC results for viral protein were consistently negative, thus lytic replication was excluded in the present study. Mild inflammation could be detected in all neural tissue samples and inflammatory infiltrates mainly consisted of CD3+ T-lymphocytes (T-cells), frequently localized in close proximity to neuronal cell bodies. To identify latently infected cell types, in situ hybridization (ISH, RNAScope®) detecting viral DNA was used on selected qPCR- positive neural tissue sections. In ganglia 30 dpi, EHV-1 ISH signal was located in the neurons of TG and ST, but also in non-neuronal support or interstitial cells surrounding the neuron. In contrast, distinct EHV-1 signal could only be observed in neurons of TG 70 dpi. Overall, detection of latent EHV-1 in abdominal tissue samples and non-neuronal cell localization suggests, that EHV-1 uses T-cells during viremia as alternative route toward latency locations in addition to retrograde neuronal transport. We therefore hypothesize that EHV-1 follows the same latency pathways as its close relative human pathogen Varicella Zoster Virus.
Keywords: Alphaherpesviruses; EHV-1; horses; latency; lymphocytes; pathogenesis; trigeminal ganglia.
Copyright © 2020 Giessler, Samoilowa, Soboll Hussey, Kiupel, Matiasek, Sledge, Liesche, Schlegel, Fux and Goehring.
Figures


Similar articles
-
Equid herpesvirus-1 Distribution in Equine Lymphoid and Neural Tissues 70 Days Post Infection.Pathogens. 2021 Jun 5;10(6):707. doi: 10.3390/pathogens10060707. Pathogens. 2021. PMID: 34198884 Free PMC article.
-
Virological and serological investigation of Equid herpesvirus 1 infection in New Zealand.Vet Microbiol. 2015 Apr 17;176(3-4):219-28. doi: 10.1016/j.vetmic.2015.01.016. Epub 2015 Jan 24. Vet Microbiol. 2015. PMID: 25666453
-
Respiratory and neurological disease in rabbits experimentally infected with equid herpesvirus 1.Microb Pathog. 2015 Oct;87:45-50. doi: 10.1016/j.micpath.2015.07.007. Epub 2015 Jul 14. Microb Pathog. 2015. PMID: 26187161
-
Equine herpesviruses 1 (EHV-1) and 4 (EHV-4)--epidemiology, disease and immunoprophylaxis: a brief review.Vet J. 2005 Jul;170(1):14-23. doi: 10.1016/j.tvjl.2004.04.018. Vet J. 2005. PMID: 15993786 Review.
-
Comparison of the pathogenesis of acute equine herpesvirus 1 (EHV-1) infection in the horse and the mouse model: a review.Vet Microbiol. 1999 Aug 16;68(1-2):3-13. doi: 10.1016/s0378-1135(99)00056-5. Vet Microbiol. 1999. PMID: 10501157 Review.
Cited by
-
Equid herpesvirus-1 Distribution in Equine Lymphoid and Neural Tissues 70 Days Post Infection.Pathogens. 2021 Jun 5;10(6):707. doi: 10.3390/pathogens10060707. Pathogens. 2021. PMID: 34198884 Free PMC article.
-
Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses.Viruses. 2021 Feb 24;13(3):356. doi: 10.3390/v13030356. Viruses. 2021. PMID: 33668216 Free PMC article.
-
The Pathogenesis and Immune Evasive Mechanisms of Equine Herpesvirus Type 1.Front Microbiol. 2021 Mar 4;12:662686. doi: 10.3389/fmicb.2021.662686. eCollection 2021. Front Microbiol. 2021. PMID: 33746936 Free PMC article. Review.
-
Attenuation of the neuropathogenic equine herpesvirus type 1 strain Ab4p in hamsters by a single amino acid mutation (D752N) in viral DNA polymerase ORF30.J Vet Med Sci. 2024 Dec 1;86(12):1273-1278. doi: 10.1292/jvms.24-0338. Epub 2024 Oct 9. J Vet Med Sci. 2024. PMID: 39384384 Free PMC article.
-
Transcriptomic Profiling of Equine and Viral Genes in Peripheral Blood Mononuclear Cells in Horses during Equine Herpesvirus 1 Infection.Pathogens. 2021 Jan 7;10(1):43. doi: 10.3390/pathogens10010043. Pathogens. 2021. PMID: 33430330 Free PMC article.
References
-
- Slater J. Equine herpesviruses. In: Sellon DC, and Long MT, editor. Equine Infectious Diseases. St. Louis, MO: Elsevier; (2014). p. 151-168.e158.
LinkOut - more resources
Full Text Sources
Miscellaneous

