Screening and Functional Prediction of Key Candidate Genes in Hepatitis B Virus-Associated Hepatocellular Carcinoma
- PMID: 33102593
- PMCID: PMC7568806
- DOI: 10.1155/2020/7653506
Screening and Functional Prediction of Key Candidate Genes in Hepatitis B Virus-Associated Hepatocellular Carcinoma
Abstract
Background: The molecular mechanism by which hepatitis B virus (HBV) induces hepatocellular carcinoma (HCC) is still unknown. The genomic expression profile and bioinformatics methods were used to investigate the potential pathogenesis and therapeutic targets for HBV-associated HCC (HBV-HCC).
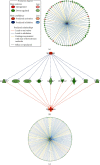
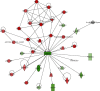
Methods: The microarray dataset GSE55092 was downloaded from the Gene Expression Omnibus (GEO) database. The data was analyzed by the bioinformatics software to find differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, ingenuity pathway analysis (IPA), and protein-protein interaction (PPI) network analysis were then performed on DEGs. The hub genes were identified using Centiscape2.2 and Molecular Complex Detection (MCODE) in the Cytoscape software (Cytoscape_v3.7.2). The survival data of these hub genes was downloaded from the Gene Expression Profiling Interactive Analysis (GEPIA).
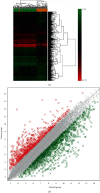
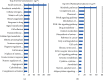
Results: A total of 2264 mRNA transcripts were differentially expressed, including 764 upregulated and 1500 downregulated in tumor tissues. GO analysis revealed that these DEGs were related to the small-molecule metabolic process, xenobiotic metabolic process, and cellular nitrogen compound metabolic process. KEGG pathway analysis revealed that metabolic pathways, complement and coagulation cascades, and chemical carcinogenesis were involved. Diseases and biofunctions showed that DEGs were mainly associated with the following diseases or biological function abnormalities: cancer, organismal injury and abnormalities, gastrointestinal disease, and hepatic system disease. The top 10 upstream regulators were predicted to be activated or inhibited by Z-score and identified 25 networks. The 10 genes with the highest degree of connectivity were defined as the hub genes. Cox regression revealed that all the 10 genes (CDC20, BUB1B, KIF11, TTK, EZH2, ZWINT, NDC80, TPX2, MELK, and KIF20A) were related to the overall survival.
Conclusion: Our study provided a registry of genes that play important roles in regulating the development of HBV-HCC, assisting us in understanding the molecular mechanisms that underlie the carcinogenesis and progression of HCC.
Copyright © 2020 Xia Chen et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

