Data on rumen and faeces microbiota profiles of Yakutian and Kalmyk cattle revealed by high-throughput sequencing of 16S rRNA gene amplicons
- PMID: 33102664
- PMCID: PMC7578675
- DOI: 10.1016/j.dib.2020.106407
Data on rumen and faeces microbiota profiles of Yakutian and Kalmyk cattle revealed by high-throughput sequencing of 16S rRNA gene amplicons
Abstract
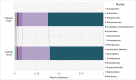
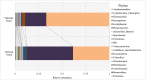
It is known that the rumen microbiome directly or indirectly contributes to animal production, and may be a prospective target for mitigation of greenhouse gas emissions [1]. At the same time, feed types and components of diet can influence the composition of the rumen microbiome [2,3]. Fluctuations in the composition of the digestive tract microbiota can alter the development, health, and productivity of cattle [4]. Many studies of cattle microbiomes have focussed on the rumen microbiota, whereas the faecal microbiota has received less attention [5], [6], [7]. Therefore, the features of the faecal and the ruminal microbiomes in different cattle breeds are yet to be studied. Here, we provided 16S rRNA gene amplicon data of the ruminal and the faecal microbiomes from Yakutian and Kalmyk cattle living in the Republic of Sakha, Yakutia, Russia. Total DNA was extracted from 13 faecal and 13 ruminal samples, and DNA libraries were prepared and sequenced on an Illumina MiSeq platform. Paired-end raw reads were processed, and final operational taxonomic units (OTUs) were assigned to the respective prokaryotic taxa using the RDP (Ribosomal Database Project) database. Analysis of the microbiome composition at the phylum level revealed very similar faecal microbiota between the introduced Kalmyk breed and the indigenous Yakutian breed, whereas the ruminal microbiomes of these breeds differed substantially in terms of relative abundance of some prokaryotic phyla. We believe that the data obtained may provide new insights into the dynamics of the ruminal and the faecal microbiota of cattle as well as disclose breed-specific features of ruminal microbiomes. Besides, these data will contribute to our understanding of the ruminal microbiome structure and function, and might be useful for the management of cattle feeding and ruminal methane production.
Keywords: 16S rRNA gene; Cattle; Faeces microbiota; Kalmyk breed; Microbiome; NGS; Rumen microbiota; Yakutian breed.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Belanche A., Doreau M., Edwards J.E., Moorby J.M., Pinloche E., Newbold C.J. Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J. Nutr. 2012;142(9):1684–1692. doi: 10.3945/jn.112.159574. - DOI - PubMed
-
- Tapio I., Fischer D., Blasco L., Tapio M., Wallace R.J., Bayat A.R., Ventto L., Kahala M., Negussie E., Shingfield K.J., Vilkki J. Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0180260. - DOI - PMC - PubMed
-
- Andrade B.G.N., Bressani F.A., Cuadrat R.R.C., Tizioto P.C., de Oliveira P.S.N., Mourão G.B., Coutinho L.L., Reecy J.M., Koltes J.E., Walsh P., Berndt A., Palhares J.C.P., Regitano L.C.A. The structure of microbial populations in Nelore GIT reveals inter-dependency of methanogens in feces and rumen. J. Anim. Sci. Biotechnol. 2020;11(6) doi: 10.1186/s40104-019-0422-x. - DOI - PMC - PubMed
-
- Dowd S.E., Callaway T.R., Wolcott R.D., Sun Y., McKeehan T., Hagevoort R.G., Edrington T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8(125) doi: 10.1186/1471-2180-8-125. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

