Optimizing open data to support one health: best practices to ensure interoperability of genomic data from bacterial pathogens
- PMID: 33103064
- PMCID: PMC7568946
- DOI: 10.1186/s42522-020-00026-3
Optimizing open data to support one health: best practices to ensure interoperability of genomic data from bacterial pathogens
Abstract
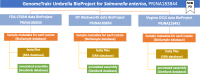
The holistic approach of One Health, which sees human, animal, plant, and environmental health as a unit, rather than discrete parts, requires not only interdisciplinary cooperation, but standardized methods for communicating and archiving data, enabling participants to easily share what they have learned and allow others to build upon their findings. Ongoing work by NCBI and the GenomeTrakr project illustrates how open data platforms can help meet the needs of federal and state regulators, public health laboratories, departments of agriculture, and universities. Here we describe how microbial pathogen surveillance can be transformed by having an open access database along with Best Practices for contributors to follow. First, we describe the open pathogen surveillance framework, hosted on the NCBI platform. We cover the current community standards for WGS quality, provide an SOP for assessing your own sequence quality and recommend QC thresholds for all submitters to follow. We then provide an overview of NCBI data submission along with step by step details. And finally, we provide curation guidance and an SOP for keeping your public data current within the database. These Best Practices can be models for other open data projects, thereby advancing the One Health goals of Findable, Accessible, Interoperable and Re-usable (FAIR) data.
Keywords: GenomeTrakr; Genomic epidemiology; Microbial pathogen surveillance; NCBI submission; One health; QA/QC; Whole genome sequencing.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
