Partial Replacement of Nucleosomal DNA with Human FACT Induces Dynamic Exposure and Acetylation of Histone H3 N-Terminal Tails
- PMID: 33103079
- PMCID: PMC7569332
- DOI: 10.1016/j.isci.2020.101641
Partial Replacement of Nucleosomal DNA with Human FACT Induces Dynamic Exposure and Acetylation of Histone H3 N-Terminal Tails
Abstract
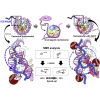
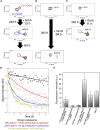
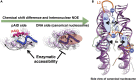
The FACT (facilitates chromatin transcription) complex, comprising SPT16 and SSRP1, conducts structural alterations during nucleosome unwrapping. Our previous cryoelectron microscopic (cryo-EM) analysis revealed the first intermediate structure of an unwrapped nucleosome with human FACT, in which 112-bp DNA and the phosphorylated intrinsically disordered (pAID) segment of SPT16 jointly wrapped around the histone core instead of 145-bp DNA. Using NMR, here we clarified that the histone H3 N-terminal tails, unobserved in the cryo-EM structure, adopt two different conformations reflecting their asymmetric locations at entry/exit sites: one corresponds to the original nucleosome site buried in two DNA gyres (DNA side), whereas the other, comprising pAID and DNA, is more exposed to the solvent (pAID side). NMR real-time monitoring showed that H3 acetylation is faster on the pAID side than on the DNA side. Our findings highlight that accessible conformations of H3 tails are created by the replacement of nucleosomal DNA with pAID.
Keywords: Biochemistry; Molecular Biology; Structural Biology.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
FACT modulates the conformations of histone H2A and H2B N-terminal tails within nucleosomes.Commun Biol. 2022 Aug 13;5(1):814. doi: 10.1038/s42003-022-03785-z. Commun Biol. 2022. PMID: 35963897 Free PMC article.
-
The N-terminal Tails of Histones H2A and H2B Adopt Two Distinct Conformations in the Nucleosome with Contact and Reduced Contact to DNA.J Mol Biol. 2021 Jul 23;433(15):167110. doi: 10.1016/j.jmb.2021.167110. Epub 2021 Jun 18. J Mol Biol. 2021. PMID: 34153285
-
Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy.Cancers (Basel). 2017 Jan 6;9(1):3. doi: 10.3390/cancers9010003. Cancers (Basel). 2017. PMID: 28067802 Free PMC article.
-
Secondary structures of the core histone N-terminal tails: their role in regulating chromatin structure.Subcell Biochem. 2013;61:37-55. doi: 10.1007/978-94-007-4525-4_2. Subcell Biochem. 2013. PMID: 23150245 Review.
-
Novel types and sites of histone modifications emerge as players in the transcriptional regulation contest.FEBS J. 2015 May;282(9):1658-74. doi: 10.1111/febs.13047. Epub 2014 Oct 7. FEBS J. 2015. PMID: 25220185 Review.
Cited by
-
Nucleosome composition regulates the histone H3 tail conformational ensemble and accessibility.Nucleic Acids Res. 2021 May 7;49(8):4750-4767. doi: 10.1093/nar/gkab246. Nucleic Acids Res. 2021. PMID: 33856458 Free PMC article.
-
The histone chaperone FACT: a guardian of chromatin structure integrity.Transcription. 2022 Feb-Jun;13(1-3):16-38. doi: 10.1080/21541264.2022.2069995. Epub 2022 Apr 29. Transcription. 2022. PMID: 35485711 Free PMC article. Review.
-
Arginine anchor points govern H3 tail dynamics.Front Mol Biosci. 2023 May 2;10:1150400. doi: 10.3389/fmolb.2023.1150400. eCollection 2023. Front Mol Biosci. 2023. PMID: 37261328 Free PMC article.
-
FACT modulates the conformations of histone H2A and H2B N-terminal tails within nucleosomes.Commun Biol. 2022 Aug 13;5(1):814. doi: 10.1038/s42003-022-03785-z. Commun Biol. 2022. PMID: 35963897 Free PMC article.
-
Drugging the "Undruggable" MYCN Oncogenic Transcription Factor: Overcoming Previous Obstacles to Impact Childhood Cancers.Cancer Res. 2021 Apr 1;81(7):1627-1632. doi: 10.1158/0008-5472.CAN-20-3108. Epub 2021 Jan 28. Cancer Res. 2021. PMID: 33509943 Free PMC article. Review.
References
-
- Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. - PubMed
LinkOut - more resources
Full Text Sources

