The mouse Gene Expression Database (GXD): 2021 update
- PMID: 33104772
- PMCID: PMC7778941
- DOI: 10.1093/nar/gkaa914
The mouse Gene Expression Database (GXD): 2021 update
Abstract
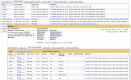
The Gene Expression Database (GXD; www.informatics.jax.org/expression.shtml) is an extensive and well-curated community resource of mouse developmental gene expression information. For many years, GXD has collected and integrated data from RNA in situ hybridization, immunohistochemistry, RT-PCR, northern blot, and western blot experiments through curation of the scientific literature and by collaborations with large-scale expression projects. Since our last report in 2019, we have continued to acquire these classical types of expression data; developed a searchable index of RNA-Seq and microarray experiments that allows users to quickly and reliably find specific mouse expression studies in ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and GEO (https://www.ncbi.nlm.nih.gov/geo/); and expanded GXD to include RNA-Seq data. Uniformly processed RNA-Seq data are imported from the EBI Expression Atlas and then integrated with the other types of expression data in GXD, and with the genetic, functional, phenotypic and disease-related information in Mouse Genome Informatics (MGI). This integration has made the RNA-Seq data accessible via GXD's enhanced searching and filtering capabilities. Further, we have embedded the Morpheus heat map utility into the GXD user interface to provide additional tools for display and analysis of RNA-Seq data, including heat map visualization, sorting, filtering, hierarchical clustering, nearest neighbors analysis and visual enrichment.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Ruzicka L., Howe D.G., Ramachandran S., Toro S., Van Slyke C.E., Bradford Y.M., Eagle A., Fashena D., Frazer K., Kalita P. et al.. The Zebrafish Information Network: new support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 2019; 47:D867–D873. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

