TriTag: an integrative tool to correlate chromatin dynamics and gene expression in living cells
- PMID: 33104788
- PMCID: PMC7736787
- DOI: 10.1093/nar/gkaa906
TriTag: an integrative tool to correlate chromatin dynamics and gene expression in living cells
Erratum in
-
TriTag: an integrative tool to correlate chromatin dynamics and gene expression in living cells.Nucleic Acids Res. 2020 Dec 16;48(22):13013-13014. doi: 10.1093/nar/gkaa1170. Nucleic Acids Res. 2020. PMID: 33219689 Free PMC article. No abstract available.
Abstract
A wealth of single-cell imaging studies have contributed novel insights into chromatin organization and gene regulation. However, a comprehensive understanding of spatiotemporal gene regulation requires developing tools to combine multiple monitoring systems in a single study. Here, we report a versatile tag, termed TriTag, which integrates the functional capabilities of CRISPR-Tag (DNA labeling), MS2 aptamer (RNA imaging) and fluorescent protein (protein tracking). Using this tag, we correlate changes in chromatin dynamics with the progression of endogenous gene expression, by recording both transcriptional bursting and protein production. This strategy allows precise measurements of gene expression at single-allele resolution across the cell cycle or in response to stress. TriTag enables capturing an integrated picture of gene expression, thus providing a powerful tool to study transcriptional heterogeneity and regulation.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

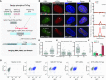
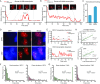


Similar articles
-
An RNA-aptamer-based two-color CRISPR labeling system.Sci Rep. 2016 May 27;6:26857. doi: 10.1038/srep26857. Sci Rep. 2016. PMID: 27229896 Free PMC article.
-
Intracellular RNA-tracking methods.Open Biol. 2018 Oct 3;8(10):180104. doi: 10.1098/rsob.180104. Open Biol. 2018. PMID: 30282659 Free PMC article. Review.
-
Aptamer-Mediated Reversible Transactivation of Gene Expression by Light.Angew Chem Int Ed Engl. 2020 Dec 7;59(50):22414-22418. doi: 10.1002/anie.202009240. Epub 2020 Oct 2. Angew Chem Int Ed Engl. 2020. PMID: 32865316 Free PMC article.
-
CRISPR-Cpf1-mediated genome editing and gene regulation in human cells.Biotechnol Adv. 2019 Jan-Feb;37(1):21-27. doi: 10.1016/j.biotechadv.2018.10.013. Epub 2018 Nov 3. Biotechnol Adv. 2019. PMID: 30399413 Review.
-
A CRISPR-Cas and Tat Peptide with Fluorescent RNA Aptamer System for Signal Amplification in RNA Imaging.Biosensors (Basel). 2023 Feb 18;13(2):293. doi: 10.3390/bios13020293. Biosensors (Basel). 2023. PMID: 36832059 Free PMC article.
Cited by
-
Research progress of live-cell RNA imaging techniques.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022 Jun 25;51(3):362-372. doi: 10.3724/zdxbyxb-2022-0017. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 36207827 Free PMC article. Review. English.
-
MONITTR allows real-time imaging of transcription and endogenous proteins in C. elegans.J Cell Biol. 2025 Jan 6;224(1):e202403198. doi: 10.1083/jcb.202403198. Epub 2024 Oct 14. J Cell Biol. 2025. PMID: 39400293 Free PMC article.
-
Gene activation guided by nascent RNA-bound transcription factors.Nat Commun. 2022 Nov 28;13(1):7329. doi: 10.1038/s41467-022-35041-7. Nat Commun. 2022. PMID: 36443367 Free PMC article.
-
CRISPR FISHer enables high-sensitivity imaging of nonrepetitive DNA in living cells through phase separation-mediated signal amplification.Cell Res. 2022 Nov;32(11):969-981. doi: 10.1038/s41422-022-00712-z. Epub 2022 Sep 14. Cell Res. 2022. PMID: 36104507 Free PMC article.
-
Illuminating the Live-Cell Dynamics of Hepatitis B Virus Covalently Closed Circular DNA Using the CRISPR-Tag System.mBio. 2023 Apr 25;14(2):e0355022. doi: 10.1128/mbio.03550-22. Epub 2023 Feb 22. mBio. 2023. PMID: 36840581 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

