Investigation of the Impact from IL-2, IL-7, and IL-15 on the Growth and Signaling of Activated CD4+ T Cells
- PMID: 33105566
- PMCID: PMC7659484
- DOI: 10.3390/ijms21217814
Investigation of the Impact from IL-2, IL-7, and IL-15 on the Growth and Signaling of Activated CD4+ T Cells
Abstract
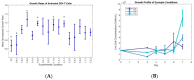
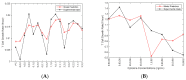
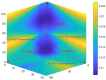
While CAR-T therapy is a growing and promising area of cancer research, it is limited by high cost and the difficulty of consistently culturing T-cells to therapeutically relevant concentrations ex-vivo. Cytokines IL-2, IL-7 and IL-15 have been found to stimulate the growth of T cells, however, the optimized combination of these three cytokines for T cell proliferation is unknown. In this study, we designed an integrated experimental and modeling approach to optimize cytokine supplementation for rapid expansion in clinical applications. We assessed the growth data for statistical improvements over no cytokine supplementation and used a systems biology approach to identify genes with the highest magnitude of expression change from control at several time points. Further, we developed a predictive mathematical model to project the growth rate for various cytokine combinations, and investigate genes and reactions regulated by cytokines in activated CD4+ T cells. The most favorable conditions from the T cell growth study and from the predictive model align to include the full range of IL-2 and IL-7 studied, and at lower levels of IL-15 (6 ng/mL or 36 ng/mL). The highest growth rates were observed where either IL-2 or IL-7 was at the highest concentration tested (15 ng/mL for IL-2 and 80 ng/mL for IL-7) while the other was at the lowest (1 ng/mL for IL-2 and 6 ng/mL for IL-7), or where both IL-2 and IL-7 concentrations are moderate-corresponding to condition keys 200, 020, and 110 respectively. This suggests a synergistic interaction of IL-2 and IL-7 with regards to promoting optimal proliferation and survival of the activated CD4+ T cells. Transcriptomic data analysis identified the genes and transcriptional regulators up/down-regulated by each of the cytokines IL-2, IL-7, and IL-15. It was found that the genes with persistent expressing changes were associated with major pathways involved in cell growth and proliferation. In addition to influencing T cell metabolism, the three cytokines were found to regulate specific genes involved in TCR, JAK/STAT, MAPK, AKT and PI3K-AKT signaling. The developed Fuzzy model that can predict the growth rate of activated CD4+ T cells for various combinations of cytokines, along with identified optimal cytokine cocktails and important genes found in transcriptomic data, can pave the way for optimizing activated CD4 T cells by regulating cytokines in the clinical setting.
Keywords: RNA sequencing; activated CD4+ T cell; interleukin-15; interleukin-2; interleukin-7.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

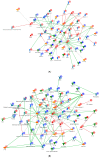

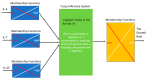
References
-
- Das K.R., Vernau L., Grupp S.A., Barrett D.M. Naive T-cell Deficits at Diagnosis and after Chemotherapy Impair Cell Therapy Potential in Pediatric Cancers. Cancer Discov. 2019;9:492–499. doi: 10.1158/2159-8290.CD-18-1314. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

