Simvastatin Induces Unfolded Protein Response and Enhances Temozolomide-Induced Cell Death in Glioblastoma Cells
- PMID: 33105603
- PMCID: PMC7690447
- DOI: 10.3390/cells9112339
Simvastatin Induces Unfolded Protein Response and Enhances Temozolomide-Induced Cell Death in Glioblastoma Cells
Erratum in
-
Correction: Dastghaib et al. Simvastatin Induces Unfolded Protein Response and Enhances Temozolomide-Induced Cell Death in Glioblastoma Cells. Cells 2020, 9, 2339.Cells. 2024 Apr 22;13(8):722. doi: 10.3390/cells13080722. Cells. 2024. PMID: 38667338 Free PMC article.
Abstract
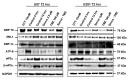
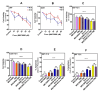
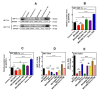
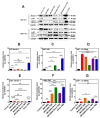
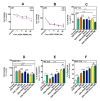
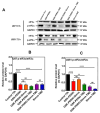
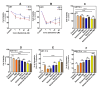
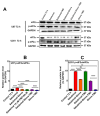
Glioblastoma (GBM) is the most prevalent malignant primary brain tumor with a very poor survival rate. Temozolomide (TMZ) is the common chemotherapeutic agent used for GBM treatment. We recently demonstrated that simvastatin (Simva) increases TMZ-induced apoptosis via the inhibition of autophagic flux in GBM cells. Considering the role of the unfolded protein response (UPR) pathway in the regulation of autophagy, we investigated the involvement of UPR in Simva-TMZ-induced cell death by utilizing highly selective IRE1 RNase activity inhibitor MKC8866, PERK inhibitor GSK-2606414 (PERKi), and eIF2α inhibitor salubrinal. Simva-TMZ treatment decreased the viability of GBM cells and significantly increased apoptotic cell death when compared to TMZ or Simva alone. Simva-TMZ induced both UPR, as determined by an increase in GRP78, XBP splicing, eukaryote initiation factor 2α (eIF2α) phosphorylation, and inhibited autophagic flux (accumulation of LC3β-II and inhibition of p62 degradation). IRE1 RNase inhibition did not affect Simva-TMZ-induced cell death, but it significantly induced p62 degradation and increased the microtubule-associated proteins light chain 3 (LC3)β-II/LC3β-I ratio in U87 cells, while salubrinal did not affect the Simva-TMZ induced cytotoxicity of GBM cells. In contrast, protein kinase RNA-like endoplasmic reticulum kinase (PERK) inhibition significantly increased Simva-TMZ-induced cell death in U87 cells. Interestingly, whereas PERK inhibition induced p62 accumulation in both GBM cell lines, it differentially affected the LC3β-II/LC3β-I ratio in U87 (decrease) and U251 (increase) cells. Simvastatin sensitizes GBM cells to TMZ-induced cell death via a mechanism that involves autophagy and UPR pathways. More specifically, our results imply that the IRE1 and PERK signaling arms of the UPR regulate Simva-TMZ-mediated autophagy flux inhibition in U251 and U87 GBM cells.
Keywords: ER stress; autophagy; autophagy flux; glioblastoma; mevalonate cascade; statin.
Conflict of interest statement
The Authors have no conflict of interest.
Figures


References
-
- Ostrom Q.T., Gittleman H., Liao P., Vecchione-Koval T., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. - DOI - PMC - PubMed
-
- Sharifzad F., Yasavoli-Sharahi H., Mardpour S., Fakharian E., Nikuinejad H., Heydari Y., Mardpour S., Taghikhani A., Khellat R., Vafaei S., et al. Neuropathological and genomic characterization of glioblastoma-induced rat model: How similar is it to humans for targeted therapy? J. Cell. Physiol. 2019;234:22493–22504. doi: 10.1002/jcp.28813. - DOI - PubMed
-
- Sharifzad F., Mardpour S., Mardpour S., Fakharian E., Taghikhani A., Sharifzad A., Kiani S., Heydarian Y., Los M.J., Azizi Z., et al. HSP70/IL-2 Treated NK Cells Effectively Cross the Blood Brain Barrier and Target Tumor Cells in a Rat Model of Induced Glioblastoma Multiforme (GBM) Int. J. Mol. Sci. 2020;21:2263. doi: 10.3390/ijms21072263. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

