Chromosome instability induced by a single defined sister chromatid fusion
- PMID: 33106324
- PMCID: PMC7652394
- DOI: 10.26508/lsa.202000911
Chromosome instability induced by a single defined sister chromatid fusion
Abstract
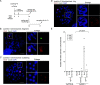
Chromosome fusion is a frequent intermediate in oncogenic chromosome rearrangements and has been proposed to cause multiple tumor-driving abnormalities. In conventional experimental systems, however, these abnormalities were often induced by randomly induced chromosome fusions involving multiple different chromosomes. It was therefore not well understood whether a single defined type of chromosome fusion, which is reminiscent of a sporadic fusion in tumor cells, has the potential to cause chromosome instabilities. Here, we developed a human cell-based sister chromatid fusion visualization system (FuVis), in which a single defined sister chromatid fusion is induced by CRISPR/Cas9 concomitantly with mCitrine expression. The fused chromosome subsequently developed extra-acentric chromosomes, including chromosome scattering, indicative of chromothripsis. Live-cell imaging and statistical modeling indicated that sister chromatid fusion generated micronuclei (MN) in the first few cell cycles and that cells with MN tend to display cell cycle abnormalities. The powerful FuVis system thus demonstrates that even a single sporadic sister chromatid fusion can induce chromosome instability and destabilize the cell cycle through MN formation.
© 2020 Kagaya et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

















Similar articles
-
53BP1 can limit sister-chromatid rupture and rearrangements driven by a distinct ultrafine DNA bridging-breakage process.Nat Commun. 2018 Feb 14;9(1):677. doi: 10.1038/s41467-018-03098-y. Nat Commun. 2018. PMID: 29445165 Free PMC article.
-
Sister chromatid cohesion defects are associated with chromosome instability in Hodgkin lymphoma cells.BMC Cancer. 2013 Aug 20;13:391. doi: 10.1186/1471-2407-13-391. BMC Cancer. 2013. PMID: 23962039 Free PMC article.
-
The number of dysfunctional telomeres in a cell: one amplifies; more than one translocate.Cytogenet Genome Res. 2008;122(3-4):315-25. doi: 10.1159/000167818. Epub 2009 Jan 30. Cytogenet Genome Res. 2008. PMID: 19188701 Review.
-
Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus.EMBO J. 2007 Feb 7;26(3):816-24. doi: 10.1038/sj.emboj.7601529. Epub 2007 Jan 25. EMBO J. 2007. PMID: 17255945 Free PMC article.
-
The molecular basis of sister-chromatid cohesion.Annu Rev Cell Dev Biol. 2001;17:753-77. doi: 10.1146/annurev.cellbio.17.1.753. Annu Rev Cell Dev Biol. 2001. PMID: 11687503 Review.
Cited by
-
Micronucleus is not a potent inducer of the cGAS/STING pathway.Life Sci Alliance. 2024 Feb 2;7(4):e202302424. doi: 10.26508/lsa.202302424. Print 2024 Apr. Life Sci Alliance. 2024. PMID: 38307626 Free PMC article.
-
Target residence of Cas9-sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing.Genome Biol. 2022 Aug 1;23(1):165. doi: 10.1186/s13059-022-02736-5. Genome Biol. 2022. PMID: 35915475 Free PMC article.
References
-
- Akaike H. (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19: 716–723. 10.1109/tac.1974.1100705 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
