Genome-wide mapping of spontaneous genetic alterations in diploid yeast cells
- PMID: 33106417
- PMCID: PMC7668089
- DOI: 10.1073/pnas.2018633117
Genome-wide mapping of spontaneous genetic alterations in diploid yeast cells
Abstract
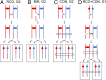
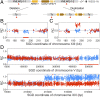
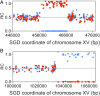
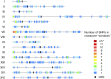
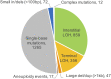
Genomic alterations including single-base mutations, deletions and duplications, translocations, mitotic recombination events, and chromosome aneuploidy generate genetic diversity. We examined the rates of all of these genetic changes in a diploid strain of Saccharomyces cerevisiae by whole-genome sequencing of many independent isolates (n = 93) subcloned about 100 times in unstressed growth conditions. The most common alterations were point mutations and small (<100 bp) insertion/deletions (n = 1,337) and mitotic recombination events (n = 1,215). The diploid cells of most eukaryotes are heterozygous for many single-nucleotide polymorphisms (SNPs). During mitotic cell divisions, recombination can produce derivatives of these cells that have become homozygous for the polymorphisms, termed loss-of-heterozygosity (LOH) events. LOH events can change the phenotype of the cells and contribute to tumor formation in humans. We observed two types of LOH events: interstitial events (conversions) resulting in a short LOH tract (usually less than 15 kb) and terminal events (mostly cross-overs) in which the LOH tract extends to the end of the chromosome. These two types of LOH events had different distributions, suggesting that they may have initiated by different mechanisms. Based on our results, we present a method of calculating the probability of an LOH event for individual SNPs located throughout the genome. We also identified several hotspots for chromosomal rearrangements (large deletions and duplications). Our results provide insights into the relative importance of different types of genetic alterations produced during vegetative growth.
Keywords: chromosome rearrangements; loss of heterozygosity; mutations; spontaneous mitotic recombination.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Hundreds of thousands of cell generations reveal a treasure chest of genome alterations.Proc Natl Acad Sci U S A. 2020 Dec 15;117(50):31567-31569. doi: 10.1073/pnas.2021185117. Epub 2020 Nov 16. Proc Natl Acad Sci U S A. 2020. PMID: 33199644 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

