Optimization of T4 phage engineering via CRISPR/Cas9
- PMID: 33106580
- PMCID: PMC7588440
- DOI: 10.1038/s41598-020-75426-6
Optimization of T4 phage engineering via CRISPR/Cas9
Abstract
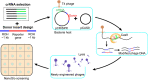
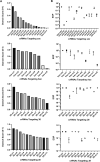
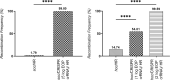
A major limitation hindering the widespread use of synthetic phages in medical and industrial settings is the lack of an efficient phage-engineering platform. Classical T4 phage engineering and several newly proposed methods are often inefficient and time consuming and consequently, only able to produce an inconsistent range of genomic editing rates between 0.03-3%. Here, we review and present new understandings of the CRISPR/Cas9 assisted genome engineering technique that significantly improves the genomic editing rate of T4 phages. Our results indicate that crRNAs selection is a major rate limiting factor in T4 phage engineering via CRISPR/Cas9. We were able to achieve an editing rate of > 99% for multiple genes that functionalizes the phages for further applications. We envision that this improved phage-engineering platform will accelerate the fields of individualized phage therapy, biocontrol, and rapid diagnostics.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

